Expression of RNautophagy/DNautophagy-related genes is regulated under control of an innate immune receptor
- PMID: 38200692
- PMCID: PMC10793664
- DOI: 10.1080/15476286.2023.2291610
Expression of RNautophagy/DNautophagy-related genes is regulated under control of an innate immune receptor
Abstract
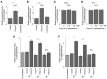
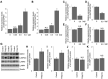
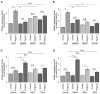
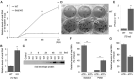
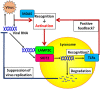
Double-stranded RNA (dsRNA) is a molecular pattern uniquely produced in cells infected with various viruses as a product or byproduct of replication. Cells detect such molecules, which indicate non-self invasion, and induce diverse immune responses to eliminate them. The degradation of virus-derived molecules can also play a role in the removal of pathogens and suppression of their replication. RNautophagy and DNautophagy are cellular degradative pathways in which RNA and DNA are directly imported into a hydrolytic organelle, the lysosome. Two lysosomal membrane proteins, SIDT2 and LAMP2C, mediate nucleic acid uptake via this pathway. Here, we showed that the expression of both SIDT2 and LAMP2C is selectively upregulated during the intracellular detection of poly(I:C), a synthetic analog of dsRNA that mimics viral infection. The upregulation of these two gene products upon poly(I:C) introduction was transient and synchronized. We also observed that the induction of SIDT2 and LAMP2C expression by poly(I:C) was dependent on MDA5, a cytoplasmic innate immune receptor that directly recognizes poly(I:C) and induces various antiviral responses. Finally, we showed that lysosomes can target viral RNA for degradation via RNautophagy and may suppress viral replication. Our results revealed a novel degradative pathway in cells as a downstream component of the innate immune response and provided evidence suggesting that the degradation of viral nucleic acids via RNautophagy/DNautophagy contributes to the suppression of viral replication.
Keywords: DNautophagy; Double-stranded RNA; Lysosome; PAMPs; PRRs; RNautophagy; immune response; innate immunity; virus infection; virus replication.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures
Similar articles
-
Cytosolic domain of SIDT2 carries an arginine-rich motif that binds to RNA/DNA and is important for the direct transport of nucleic acids into lysosomes.Autophagy. 2020 Nov;16(11):1974-1988. doi: 10.1080/15548627.2020.1712109. Epub 2020 Jan 16. Autophagy. 2020. PMID: 31944164 Free PMC article.
-
An RNautophagy/DNautophagy receptor, LAMP2C, possesses an arginine-rich motif that mediates RNA/DNA-binding.Biochem Biophys Res Commun. 2015 May 1;460(2):281-6. doi: 10.1016/j.bbrc.2015.03.025. Epub 2015 Mar 13. Biochem Biophys Res Commun. 2015. PMID: 25772617
-
Direct uptake and degradation of DNA by lysosomes.Autophagy. 2013 Aug;9(8):1167-71. doi: 10.4161/auto.24880. Epub 2013 May 21. Autophagy. 2013. PMID: 23839276 Free PMC article.
-
Strategies of highly pathogenic RNA viruses to block dsRNA detection by RIG-I-like receptors: hide, mask, hit.Antiviral Res. 2013 Dec;100(3):615-35. doi: 10.1016/j.antiviral.2013.10.002. Epub 2013 Oct 12. Antiviral Res. 2013. PMID: 24129118 Free PMC article. Review.
-
A Novel Mechanism Underlying the Innate Immune Response Induction upon Viral-Dependent Replication of Host Cell mRNA: A Mistake of +sRNA Viruses' Replicases.Front Cell Infect Microbiol. 2017 Jan 20;7:5. doi: 10.3389/fcimb.2017.00005. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28164038 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

