Processing of stalled replication forks in Bacillus subtilis
- PMID: 38052445
- PMCID: PMC10804225
- DOI: 10.1093/femsre/fuad065
Processing of stalled replication forks in Bacillus subtilis
Abstract
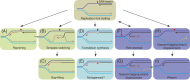
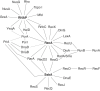
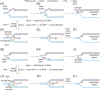
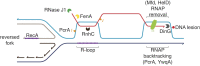
Accurate DNA replication and transcription elongation are crucial for preventing the accumulation of unreplicated DNA and genomic instability. Cells have evolved multiple mechanisms to deal with impaired replication fork progression, challenged by both intrinsic and extrinsic impediments. The bacterium Bacillus subtilis, which adopts multiple forms of differentiation and development, serves as an excellent model system for studying the pathways required to cope with replication stress to preserve genomic stability. This review focuses on the genetics, single molecule choreography, and biochemical properties of the proteins that act to circumvent the replicative arrest allowing the resumption of DNA synthesis. The RecA recombinase, its mediators (RecO, RecR, and RadA/Sms) and modulators (RecF, RecX, RarA, RecU, RecD2, and PcrA), repair licensing (DisA), fork remodelers (RuvAB, RecG, RecD2, RadA/Sms, and PriA), Holliday junction resolvase (RecU), nucleases (RnhC and DinG), and translesion synthesis DNA polymerases (PolY1 and PolY2) are key functions required to overcome a replication stress, provided that the fork does not collapse.
Keywords: DNA damage tolerance; RNA polymerase hub; RecA hub; SsbA hub; fork reversal; replisome disassembly; template switching.
© The Author(s) 2023. Published by Oxford University Press on behalf of FEMS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Bacillus subtilis DisA helps to circumvent replicative stress during spore revival.DNA Repair (Amst). 2017 Nov;59:57-68. doi: 10.1016/j.dnarep.2017.09.006. Epub 2017 Sep 22. DNA Repair (Amst). 2017. PMID: 28961460
-
Activity and in vivo dynamics of Bacillus subtilis DisA are affected by RadA/Sms and by Holliday junction-processing proteins.DNA Repair (Amst). 2017 Jul;55:17-30. doi: 10.1016/j.dnarep.2017.05.002. Epub 2017 May 5. DNA Repair (Amst). 2017. PMID: 28511132
-
Interplay between Bacillus subtilis RecD2 and the RecG or RuvAB helicase in recombinational repair.DNA Repair (Amst). 2017 Jul;55:40-46. doi: 10.1016/j.dnarep.2017.05.004. Epub 2017 May 12. DNA Repair (Amst). 2017. PMID: 28527403
-
DNA Helicase-SSB Interactions Critical to the Regression and Restart of Stalled DNA Replication forks in Escherichia coli.Genes (Basel). 2020 Apr 26;11(5):471. doi: 10.3390/genes11050471. Genes (Basel). 2020. PMID: 32357475 Free PMC article. Review.
-
Generation and Repair of Postreplication Gaps in Escherichia coli.Microbiol Mol Biol Rev. 2023 Jun 28;87(2):e0007822. doi: 10.1128/mmbr.00078-22. Epub 2023 May 22. Microbiol Mol Biol Rev. 2023. PMID: 37212693 Free PMC article. Review.
References
-
- Aliotta JM, Pelletier JJ, Ware JLet al. . Thermostable BstDNA polymerase I lacks a 3′→5′ proofreading exonuclease activity. Genet Anal Biomol Eng. 1996;12:185–95. - PubMed
-
- Alonso JC, Cárdenas PP, Sanchez Het al. . Early steps of double-strand break repair in Bacillus subtilis. DNA Repair. 2013;12:162–76. - PubMed
-
- Alonso JC, Lüder G, Trautner TA. Intramolecular homologous recombination in Bacillus subtilis 168. Molec Gen Genet. 1992;236:60–4. - PubMed
-
- Alonso JC, Lüder G. Characterization of recF suppressors in Bacillus subtilis. Biochimie. 1991;73:277–80. - PubMed


