Lola-I is a promoter pioneer factor that establishes de novo Pol II pausing during development
- PMID: 37735176
- PMCID: PMC10514308
- DOI: 10.1038/s41467-023-41408-1
Lola-I is a promoter pioneer factor that establishes de novo Pol II pausing during development
Abstract
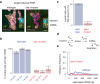
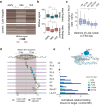
While the accessibility of enhancers is dynamically regulated during development, promoters tend to be constitutively accessible and poised for activation by paused Pol II. By studying Lola-I, a Drosophila zinc finger transcription factor, we show here that the promoter state can also be subject to developmental regulation independently of gene activation. Lola-I is ubiquitously expressed at the end of embryogenesis and causes its target promoters to become accessible and acquire paused Pol II throughout the embryo. This promoter transition is required but not sufficient for tissue-specific target gene activation. Lola-I mediates this function by depleting promoter nucleosomes, similar to the action of pioneer factors at enhancers. These results uncover a level of regulation for promoters that is normally found at enhancers and reveal a mechanism for the de novo establishment of paused Pol II at promoters.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
RNA Polymerase II "Pause" Prepares Promoters for Upcoming Transcription during Drosophila Development.Int J Mol Sci. 2022 Sep 13;23(18):10662. doi: 10.3390/ijms231810662. Int J Mol Sci. 2022. PMID: 36142573 Free PMC article.
-
Distinct mechanisms of transcriptional pausing orchestrated by GAGA factor and M1BP, a novel transcription factor.EMBO J. 2013 Jul 3;32(13):1829-41. doi: 10.1038/emboj.2013.111. Epub 2013 May 24. EMBO J. 2013. PMID: 23708796 Free PMC article.
-
GAGA factor maintains nucleosome-free regions and has a role in RNA polymerase II recruitment to promoters.PLoS Genet. 2015 Mar 27;11(3):e1005108. doi: 10.1371/journal.pgen.1005108. eCollection 2015 Mar. PLoS Genet. 2015. PMID: 25815464 Free PMC article.
-
Promoter-proximal pausing of RNA polymerase II: a nexus of gene regulation.Genes Dev. 2019 Aug 1;33(15-16):960-982. doi: 10.1101/gad.325142.119. Epub 2019 May 23. Genes Dev. 2019. PMID: 31123063 Free PMC article. Review.
-
RNA polymerase II pausing during development.Development. 2014 Mar;141(6):1179-83. doi: 10.1242/dev.088492. Development. 2014. PMID: 24595285 Free PMC article. Review.
Cited by
-
Exploring the reciprocity between pioneer factors and development.Development. 2024 Jul 1;151(13):dev201921. doi: 10.1242/dev.201921. Epub 2024 Jul 3. Development. 2024. PMID: 38958075 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases


