Identification and verification of plasma protein biomarkers that accurately identify an ectopic pregnancy
- PMID: 37715129
- PMCID: PMC10503165
- DOI: 10.1186/s12014-023-09425-w
Identification and verification of plasma protein biomarkers that accurately identify an ectopic pregnancy
Abstract
Background: Differentiating between a normal intrauterine pregnancy (IUP) and abnormal conditions including early pregnancy loss (EPL) or ectopic pregnancy (EP) is a major clinical challenge in early pregnancy. Currently, serial β-human chorionic gonadotropin (β-hCG) and progesterone are the most commonly used plasma biomarkers for evaluating pregnancy prognosis when ultrasound is inconclusive. However, neither biomarker can predict an EP with sufficient and reproducible accuracy. Hence, identification of new plasma biomarkers that can accurately diagnose EP would have great clinical value.
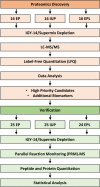
Methods: Plasma was collected from a discovery cohort of 48 consenting women having an IUP, EPL, or EP. Samples were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) followed by a label-free proteomics analysis to identify significant changes between pregnancy outcomes. A panel of 14 candidate biomarkers were then verified in an independent cohort of 74 women using absolute quantitation by targeted parallel reaction monitoring mass spectrometry (PRM-MS) which provided the capacity to distinguish between closely related protein isoforms. Logistic regression and Lasso feature selection were used to evaluate the performance of individual biomarkers and panels of multiple biomarkers to predict EP.
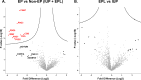
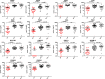
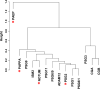
Results: A total of 1391 proteins were identified in an unbiased plasma proteome discovery. A number of significant changes (FDR ≤ 5%) were identified when comparing EP vs. non-EP (IUP + EPL). Next, 14 candidate biomarkers (ADAM12, CGA, CGB, ISM2, NOTUM, PAEP, PAPPA, PSG1, PSG2, PSG3, PSG9, PSG11, PSG6/9, and PSG8/1) were verified as being significantly different between EP and non-EP in an independent cohort (FDR ≤ 5%). Using logistic regression models, a risk score for EP was calculated for each subject, and four multiple biomarker logistic models were identified that performed similarly and had higher AUCs than models with single predictors.
Conclusions: Overall, four multivariable logistic models were identified that had significantly better prediction of having EP than those logistic models with single biomarkers. Model 4 (NOTUM, PAEP, PAPPA, ADAM12) had the highest AUC (0.987) and accuracy (96%). However, because the models are statistically similar, all markers in the four models and other highly correlated markers should be considered in further validation studies.
Keywords: Biomarker signatures; Biomarkers; Ectopic pregnancy; Miscarriage; Proteomics; Targeted MS.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
LAB, DWS, and KTB are inventors on patents for EP diagnosis. No other disclosures were reported.
Figures


Similar articles
-
Proteome-defined changes in cellular pathways for decidua and trophoblast tissues associated with location and viability of early-stage pregnancy.Reprod Biol Endocrinol. 2022 Feb 21;20(1):36. doi: 10.1186/s12958-022-00908-3. Reprod Biol Endocrinol. 2022. PMID: 35189928 Free PMC article.
-
Systematic discovery of ectopic pregnancy serum biomarkers using 3-D protein profiling coupled with label-free quantitation.J Proteome Res. 2011 Mar 4;10(3):1126-38. doi: 10.1021/pr1008866. Epub 2011 Jan 7. J Proteome Res. 2011. PMID: 21142075 Free PMC article.
-
Comparison of the diagnostic values of circulating steroid hormones, VEGF-A, PIGF, and ADAM12 in women with ectopic pregnancy.J Transl Med. 2013 Feb 19;11:44. doi: 10.1186/1479-5876-11-44. J Transl Med. 2013. PMID: 23421942 Free PMC article.
-
First trimester serum tests for Down's syndrome screening.Cochrane Database Syst Rev. 2015 Nov 30;2015(11):CD011975. doi: 10.1002/14651858.CD011975. Cochrane Database Syst Rev. 2015. PMID: 26617074 Free PMC article. Review.
-
Diagnosing ectopic pregnancy and current concepts in the management of pregnancy of unknown location.Hum Reprod Update. 2014 Mar-Apr;20(2):250-61. doi: 10.1093/humupd/dmt047. Epub 2013 Oct 6. Hum Reprod Update. 2014. PMID: 24101604 Review.
Cited by
-
NOTUM-mediated WNT silencing drives extravillous trophoblast cell lineage development.Proc Natl Acad Sci U S A. 2024 Oct;121(40):e2403003121. doi: 10.1073/pnas.2403003121. Epub 2024 Sep 26. Proc Natl Acad Sci U S A. 2024. PMID: 39325428
-
NOTUM-MEDIATED WNT SILENCING DRIVES EXTRAVILLOUS TROPHOBLAST CELL LINEAGE DEVELOPMENT.bioRxiv [Preprint]. 2024 Aug 22:2024.02.13.579974. doi: 10.1101/2024.02.13.579974. bioRxiv. 2024. Update in: Proc Natl Acad Sci U S A. 2024 Oct;121(40):e2403003121. doi: 10.1073/pnas.2403003121 PMID: 38405745 Free PMC article. Updated. Preprint.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

