TP53 germline pathogenic variants in modern humans were likely originated during recent human history
- PMID: 37304756
- PMCID: PMC10251638
- DOI: 10.1093/narcan/zcad025
TP53 germline pathogenic variants in modern humans were likely originated during recent human history
Abstract
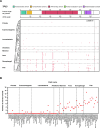
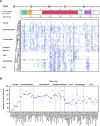
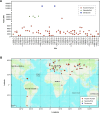
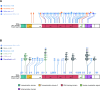
TP53 is crucial for maintaining genome stability and preventing oncogenesis. Germline pathogenic variation in TP53 damages its function, causing genome instability and increased cancer risk. Despite extensive study in TP53, the evolutionary origin of the human TP53 germline pathogenic variants remains largely unclear. In this study, we applied phylogenetic and archaeological approaches to identify the evolutionary origin of TP53 germline pathogenic variants in modern humans. In the phylogenic analysis, we searched 406 human TP53 germline pathogenic variants in 99 vertebrates distributed in eight clades of Primate, Euarchontoglires, Laurasiatheria, Afrotheria, Mammal, Aves, Sarcopterygii and Fish, but we observed no direct evidence for the cross-species conservation as the origin; in the archaeological analysis, we searched the variants in 5031 ancient human genomes dated between 45045 and 100 years before present, and identified 45 pathogenic variants in 62 ancient humans dated mostly within the last 8000 years; we also identified 6 pathogenic variants in 3 Neanderthals dated 44000 to 38515 years before present and 1 Denisovan dated 158 550 years before present. Our study reveals that TP53 germline pathogenic variants in modern humans were likely originated in recent human history and partially inherited from the extinct Neanderthals and Denisovans.
© The Author(s) 2023. Published by Oxford University Press on behalf of NAR Cancer.
Figures

Similar articles
-
Pathogenic variants in human DNA damage repair genes mostly arose in recent human history.BMC Cancer. 2024 Apr 4;24(1):415. doi: 10.1186/s12885-024-12160-6. BMC Cancer. 2024. PMID: 38575974 Free PMC article.
-
Evolutionary Origin of MUTYH Germline Pathogenic Variations in Modern Humans.Biomolecules. 2023 Feb 24;13(3):429. doi: 10.3390/biom13030429. Biomolecules. 2023. PMID: 36979362 Free PMC article.
-
Human BRCA pathogenic variants were originated during recent human history.Life Sci Alliance. 2022 Feb 14;5(5):e202101263. doi: 10.26508/lsa.202101263. Print 2022 May. Life Sci Alliance. 2022. PMID: 35165121 Free PMC article.
-
Current review of TP53 pathogenic germline variants in breast cancer patients outside Li-Fraumeni syndrome.Hum Mutat. 2018 Dec;39(12):1764-1773. doi: 10.1002/humu.23656. Epub 2018 Oct 3. Hum Mutat. 2018. PMID: 30240537 Review.
-
Radiotherapy-induced malignancies in breast cancer patients with TP53 pathogenic germline variants (Li-Fraumeni syndrome).Fam Cancer. 2020 Jan;19(1):47-53. doi: 10.1007/s10689-019-00153-5. Fam Cancer. 2020. PMID: 31748977 Review.
Cited by
-
Therapy-relevant MDM2 amplification in cholangiocarcinomas in Caucasian patients.Ther Adv Med Oncol. 2024 Nov 9;16:17588359241288123. doi: 10.1177/17588359241288123. eCollection 2024. Ther Adv Med Oncol. 2024. PMID: 39525665 Free PMC article.
-
Pathogenic variants in human DNA damage repair genes mostly arose after the latest human out-of-Africa migration.Front Genet. 2024 Jun 14;15:1408952. doi: 10.3389/fgene.2024.1408952. eCollection 2024. Front Genet. 2024. PMID: 38948361 Free PMC article.
-
Using Portuguese BRCA pathogenic variation as a model to study the impact of human admixture on human health.BMC Genomics. 2024 Apr 27;25(1):416. doi: 10.1186/s12864-024-10311-4. BMC Genomics. 2024. PMID: 38671360 Free PMC article.
-
Pathogenic variants in human DNA damage repair genes mostly arose in recent human history.BMC Cancer. 2024 Apr 4;24(1):415. doi: 10.1186/s12885-024-12160-6. BMC Cancer. 2024. PMID: 38575974 Free PMC article.
-
Onco-Breastomics: An Eco-Evo-Devo Holistic Approach.Int J Mol Sci. 2024 Jan 28;25(3):1628. doi: 10.3390/ijms25031628. Int J Mol Sci. 2024. PMID: 38338903 Free PMC article. Review.
References
-
- Levine A.J. p53: 800 million years of evolution and 40 years of discovery. Nat. Rev. Cancer. 2020; 20:471–480. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous


