Stacking Multiple Genes Improves Resistance to Chilo suppressalis, Magnaporthe oryzae, and Nilaparvata lugens in Transgenic Rice
- PMID: 37239430
- PMCID: PMC10218540
- DOI: 10.3390/genes14051070
Stacking Multiple Genes Improves Resistance to Chilo suppressalis, Magnaporthe oryzae, and Nilaparvata lugens in Transgenic Rice
Abstract
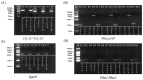
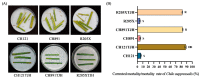
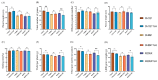
The ability of various pests and diseases to adapt to a single plant resistance gene over time leads to loss of resistance in transgenic rice. Therefore, introduction of different pest and disease resistance genes is critical for successful cultivation of transgenic rice strains with broad-spectrum resistance to multiple pathogens. Here, we produced resistance rice lines with multiple, stacked resistance genes by stacking breeding and comprehensively evaluated their resistance to Chilo suppressalis (striped rice stemborer), Magnaporthe oryzae (rice blast), and Nilaparvata lugens (brown planthopper) in a pesticide-free environment. CRY1C and CRY2A are exogenous genes from Bacillus thuringiensis. Pib, Pikm, and Bph29 are natural genes in rice. CH121TJH was introduced into CRY 1C, Pib, Pikm, and Bph29. CH891TJH and R205XTJH were introduced into CRY 2A, Pib, Pikm, and Bph29. Compared with those observed in their recurrent parents, CH121TJH significantly increased the mortality of borers. The other two lines CH891TJH and R205XTJH are the same result. Three lines introduction of Pib and Pikm significantly reduced the area of rice blast lesions, and introduction of Bph29 significantly reduced seedling mortality from N. lugens. Introduction of the exogenous genes had relatively few effects on agronomic and yield traits of the original parents. These findings suggest that stacking of rice resistance genes through molecular marker-assisted backcross breeding can confer broad spectrum and multiple resistance in differently genetic backgrounds.
Keywords: Chilo suppressalis; Magnaporthe oryzae; Nilaparvata lugens; rice blast; transgenic rice.
Conflict of interest statement
All authors declare that they have no conflict of interest.
Figures




Similar articles
-
Downregulation of Chilo suppressalis alkaline phosphatase genes associated with resistance to three transgenic Bacillus thuringiensis rice lines.Insect Mol Biol. 2018 Feb;27(1):83-89. doi: 10.1111/imb.12349. Epub 2017 Sep 21. Insect Mol Biol. 2018. PMID: 28940938
-
Cloning and functional identification of a Chilo suppressalis-inducible promoter of rice gene, OsHPL2.Pest Manag Sci. 2020 Sep;76(9):3177-3187. doi: 10.1002/ps.5872. Epub 2020 May 10. Pest Manag Sci. 2020. PMID: 32336018
-
Co-transformation mediated stacking of blast resistance genes Pi54 and Pi54rh in rice provides broad spectrum resistance against Magnaporthe oryzae.Plant Cell Rep. 2017 Nov;36(11):1747-1755. doi: 10.1007/s00299-017-2189-x. Epub 2017 Sep 13. Plant Cell Rep. 2017. PMID: 28905253
-
Recent progress on the genetics and molecular breeding of brown planthopper resistance in rice.Rice (N Y). 2016 Dec;9(1):30. doi: 10.1186/s12284-016-0099-0. Epub 2016 Jun 14. Rice (N Y). 2016. PMID: 27300326 Free PMC article. Review.
-
Defense Strategies of Rice in Response to the Attack of the Herbivorous Insect, Chilo suppressalis.Int J Mol Sci. 2023 Sep 21;24(18):14361. doi: 10.3390/ijms241814361. Int J Mol Sci. 2023. PMID: 37762665 Free PMC article. Review.
References
-
- Litsinger J.A., Canapi B.L., Bandong J.P., Cruz C.D., Apostol R.F., Pantua P.C., Lumaban M.D., Alviola A.L., Raymundo F., Libetario E.M., et al. Rice Crop Loss from Insect Pests in Wetland and Dryland Environments of Asia with Emphasis on the Philippines. Int. J. Trop. Insect Sci. 2011;8:677–692. doi: 10.1017/S1742758400022785. - DOI
-
- Jiang Y., Pan S., Cai M., Li C., Zhan M., Wang J., Mohamed I., Cao C. Assessment of yield advantages of Bt-MH63 with cry1C* or cry2A* genes over MH63 (Oryza sativa L.) under different pest control modes. Field Crops Res. 2014;155:153–158. doi: 10.1016/j.fcr.2013.09.011. - DOI
-
- Khush G.S., Jena K.K. Advances in Genetics, Genomics and Control of Rice Blast Disease. Conf. Proc. 2009;1:1–10. doi: 10.1007/978-1-4020-9500-9_1. - DOI
-
- Pham V.D., Cabunagan R.C., Cabauatan P.Q., Choi H.S., Choi I.R., Ho V.C., Nguyen H.H. Yellowing syndrome of rice: Etiology, current status and future challenges. Omonrice. 2007;15:94–101.
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials


