Characterization of Two Transposable Elements and an Ultra-Conserved Element Isolated in the Genome of Zootoca vivipara (Squamata, Lacertidae)
- PMID: 36983793
- PMCID: PMC10058329
- DOI: 10.3390/life13030637
Characterization of Two Transposable Elements and an Ultra-Conserved Element Isolated in the Genome of Zootoca vivipara (Squamata, Lacertidae)
Abstract
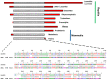
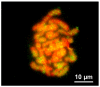
Transposable elements (TEs) constitute a considerable fraction of eukaryote genomes representing a major source of genetic variability. We describe two DNA sequences isolated in the lizard Zootoca vivipara, here named Zv516 and Zv817. Both sequences are single-copy nuclear sequences, including a truncation of two transposable elements (TEs), SINE Squam1 in Zv516 and a Tc1/Mariner-like DNA transposon in Zv817. FISH analyses with Zv516 showed the occurrence of interspersed signals of the SINE Squam1 sequence on all chromosomes of Z. vivipara and quantitative dot blot indicated that this TE is present with about 4700 copies in the Z. vivipara genome. FISH and dot blot with Zv817 did not produce clear hybridization signals. Bioinformatic analysis showed the presence of active SINE Squam 1 copies in the genome of different lacertids, in different mRNAs, and intronic and coding regions of various genes. The Tc1/Mariner-like DNA transposon occurs in all reptiles, excluding Sphenodon and Archosauria. Zv817 includes a trait of 284 bp, representing an amniote ultra-conserved element (UCE). Using amniote UCE homologous sequences from available whole genome sequences of major amniote taxonomic groups, we performed a phylogenetic analysis which retrieved Prototheria as the sister group of Metatheria and Eutheria. Within diapsids, Testudines are the sister group to Aves + Crocodylia (Archosauria), and Sphenodon is the sister group to Squamata. Furthermore, large trait regions flanking the UCE are conserved at family level.
Keywords: DNA transposons; SINEs; Tc1/Mariner; amniotes; evolutionarily conserved elements; reptiles; squamates.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Isolation and Characterization of Interspersed Repeated Sequences in the Common Lizard, Zootoca vivipara, and Their Conservation in Squamata.Cytogenet Genome Res. 2019;157(1-2):65-76. doi: 10.1159/000497304. Epub 2019 Mar 6. Cytogenet Genome Res. 2019. PMID: 30836364
-
Characterization of a Tc1-like transposable element in zebrafish (Danio rerio).Mol Gen Genet. 1995 May 10;247(3):312-22. doi: 10.1007/BF00293199. Mol Gen Genet. 1995. PMID: 7770036
-
Short interspersed elements (SINEs) of squamate reptiles (Squam1 and Squam2): structure and phylogenetic significance.J Exp Zool B Mol Dev Evol. 2011 May 15;316B(3):212-26. doi: 10.1002/jez.b.21391. Epub 2010 Dec 31. J Exp Zool B Mol Dev Evol. 2011. PMID: 21462315
-
A Brief Review of Meiotic Chromosomes in Early Spermatogenesis and Oogenesis and Mitotic Chromosomes in the Viviparous Lizard Zootoca vivipara (Squamata: Lacertidae) with Multiple Sex Chromosomes.Animals (Basel). 2022 Dec 20;13(1):19. doi: 10.3390/ani13010019. Animals (Basel). 2022. PMID: 36611629 Free PMC article. Review.
-
Human transposable elements in Repbase: genomic footprints from fish to humans.Mob DNA. 2018 Jan 4;9:2. doi: 10.1186/s13100-017-0107-y. eCollection 2018. Mob DNA. 2018. PMID: 29308093 Free PMC article. Review.
Cited by
-
The Satellite DNA PcH-Sat, Isolated and Characterized in the Limpet Patella caerulea (Mollusca, Gastropoda), Suggests the Origin from a Nin-SINE Transposable Element.Genes (Basel). 2024 Apr 25;15(5):541. doi: 10.3390/genes15050541. Genes (Basel). 2024. PMID: 38790169 Free PMC article.
References
-
- Kidwell M.G., Lisch D.R. Perspective: Transposable elements, parasitic DNA, and genome evolution. Evolution. 2001;55:1–24. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources


