Evolutionary Origin of MUTYH Germline Pathogenic Variations in Modern Humans
- PMID: 36979362
- PMCID: PMC10046817
- DOI: 10.3390/biom13030429
Evolutionary Origin of MUTYH Germline Pathogenic Variations in Modern Humans
Abstract
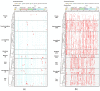
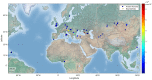
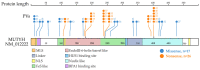
MUTYH plays an essential role in preventing oxidation-caused DNA damage. Pathogenic germline variations in MUTYH damage its function, causing intestinal polyposis and colorectal cancer. Determination of the evolutionary origin of the variation is essential to understanding the etiological relationship between MUTYH variation and cancer development. In this study, we analyzed the origins of pathogenic germline variants in human MUTYH. Using a phylogenic approach, we searched MUTYH pathogenic variants in modern humans in the MUTYH of 99 vertebrates across eight clades. We did not find pathogenic variants shared between modern humans and the non-human vertebrates following the evolutionary tree, ruling out the possibility of cross-species conservation as the origin of human pathogenic variants in MUTYH. We then searched the variants in the MUTYH of 5031 ancient humans and extinct Neanderthals and Denisovans. We identified 24 pathogenic variants in 42 ancient humans dated between 30,570 and 480 years before present (BP), and three pathogenic variants in Neanderthals dated between 65,000 and 38,310 years BP. Data from our study revealed that human MUTYH pathogenic variants mostly arose in recent human history and partially originated from Neanderthals.
Keywords: MUTYH; ancient humans; archaeological; evolutionary origin; pathogenic variant; phylogenic.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
TP53 germline pathogenic variants in modern humans were likely originated during recent human history.NAR Cancer. 2023 Jun 9;5(3):zcad025. doi: 10.1093/narcan/zcad025. eCollection 2023 Sep. NAR Cancer. 2023. PMID: 37304756 Free PMC article.
-
Low frequency of AXIN2 mutations and high frequency of MUTYH mutations in patients with multiple polyposis.Hum Mutat. 2006 Oct;27(10):1064. doi: 10.1002/humu.9460. Hum Mutat. 2006. PMID: 16941501
-
A comprehensive characterization of the spectrum of MUTYH germline pathogenic variants in Latin America.Fam Cancer. 2024 Nov;23(4):507-513. doi: 10.1007/s10689-024-00382-3. Epub 2024 Apr 30. Fam Cancer. 2024. PMID: 38687439
-
[From gene to disease; MutYH-associated polyposis coli (MAP)].Ned Tijdschr Geneeskd. 2005 Dec 31;149(53):2970-2. Ned Tijdschr Geneeskd. 2005. PMID: 16425850 Review. Dutch.
-
MUTYH-associated colorectal cancer and adenomatous polyposis.Surg Today. 2014 Apr;44(4):593-600. doi: 10.1007/s00595-013-0592-7. Epub 2013 Apr 19. Surg Today. 2014. PMID: 23605219 Review.
Cited by
-
Pathogenic variants in human DNA damage repair genes mostly arose after the latest human out-of-Africa migration.Front Genet. 2024 Jun 14;15:1408952. doi: 10.3389/fgene.2024.1408952. eCollection 2024. Front Genet. 2024. PMID: 38948361 Free PMC article.
-
Pathogenic variants in human DNA damage repair genes mostly arose in recent human history.BMC Cancer. 2024 Apr 4;24(1):415. doi: 10.1186/s12885-024-12160-6. BMC Cancer. 2024. PMID: 38575974 Free PMC article.
-
Classification of MLH1 Missense VUS Using Protein Structure-Based Deep Learning-Ramachandran Plot-Molecular Dynamics Simulations Method.Int J Mol Sci. 2024 Jan 10;25(2):850. doi: 10.3390/ijms25020850. Int J Mol Sci. 2024. PMID: 38255924 Free PMC article.
References
-
- Slupska M.M., Baikalov C., Luther W.M., Chiang J.H., Wei Y.F., Miller J.H. Cloning and Sequencing a Human Homolog (HMYH) of the Escherichia Coli MutY Gene Whose Function Is Required for the Repair of Oxidative DNA Damage. J. Bacteriol. 1996;178:3885–3892. doi: 10.1128/jb.178.13.3885-3892.1996. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials


