Diverse viromes in polar regions: A retrospective study of metagenomic data from Antarctic animal feces and Arctic frozen soil in 2012-2014
- PMID: 36028202
- PMCID: PMC9797369
- DOI: 10.1016/j.virs.2022.08.006
Diverse viromes in polar regions: A retrospective study of metagenomic data from Antarctic animal feces and Arctic frozen soil in 2012-2014
Abstract
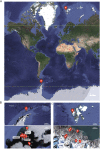
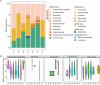
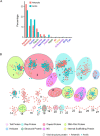
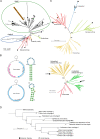
Antarctica and the Arctic are the coldest places, containing a high diversity of microorganisms, including viruses, which are important components of polar ecosystems. However, owing to the difficulties in obtaining access to animal and environmental samples, the current knowledge of viromes in polar regions is still limited. To better understand polar viromes, this study performed a retrospective analysis using metagenomic sequencing data of animal feces from Antarctica and frozen soil from the Arctic collected during 2012-2014. The results reveal diverse communities of DNA and RNA viruses from at least 23 families from Antarctic animal feces and 16 families from Arctic soils. Although the viral communities from Antarctica and the Arctic show a large diversity, they have genetic similarities with known viruses from different ecosystems and organisms with similar viral proteins. Phylogenetic analysis of Microviridae, Parvoviridae, and Larvidaviridae was further performed, and complete genomic sequences of two novel circular replication-associated protein (rep)-encoding single-stranded (CRESS) DNA viruses closely related to Circoviridae were identified. These results reveal the high diversity, complexity, and novelty of viral communities from polar regions, and suggested the genetic similarity and functional correlations of viromes between the Antarctica and Arctic. Variations in viral families in Arctic soils, Arctic freshwater, and Antarctic soils are discussed. These findings improve our understanding of polar viromes and suggest the importance of performing follow-up in-depth investigations of animal and environmental samples from Antarctica and the Arctic, which would reveal the substantial role of these viruses in the global viral community.
Keywords: Antarctica animal feces; Arctic soil; Metagenomics; Virus diversity.
Copyright © 2022 The Authors. Publishing services by Elsevier B.V. All rights reserved.
Figures

Similar articles
-
Viruses in Polar Lake and Soil Ecosystems.Adv Virus Res. 2018;101:39-54. doi: 10.1016/bs.aivir.2018.02.002. Epub 2018 Apr 26. Adv Virus Res. 2018. PMID: 29908593 Review.
-
Single-Stranded DNA Viruses in Antarctic Cryoconite Holes.Viruses. 2019 Nov 4;11(11):1022. doi: 10.3390/v11111022. Viruses. 2019. PMID: 31689942 Free PMC article.
-
Biodiversity and distribution of polar freshwater DNA viruses.Sci Adv. 2015 Jun 19;1(5):e1400127. doi: 10.1126/sciadv.1400127. eCollection 2015 Jun. Sci Adv. 2015. PMID: 26601189 Free PMC article.
-
Diversity and Distribution Characteristics of Viruses in Soils of a Marine-Terrestrial Ecotone in East China.Microb Ecol. 2018 Feb;75(2):375-386. doi: 10.1007/s00248-017-1049-0. Epub 2017 Aug 21. Microb Ecol. 2018. PMID: 28825127
-
Viruses of Polar Aquatic Environments.Viruses. 2019 Feb 22;11(2):189. doi: 10.3390/v11020189. Viruses. 2019. PMID: 30813316 Free PMC article. Review.
Cited by
-
An overview for monitoring and prediction of pathogenic microorganisms in the atmosphere.Fundam Res. 2023 Aug 28;4(3):430-441. doi: 10.1016/j.fmre.2023.05.022. eCollection 2024 May. Fundam Res. 2023. PMID: 38933199 Free PMC article. Review.
-
Breaking the Ice: A Review of Phages in Polar Ecosystems.Methods Mol Biol. 2024;2738:31-71. doi: 10.1007/978-1-0716-3549-0_3. Methods Mol Biol. 2024. PMID: 37966591 Review.
-
A Pseudomonas Lysogenic Bacteriophage Crossing the Antarctic and Arctic, Representing a New Genus of Autographiviridae.Int J Mol Sci. 2023 Apr 21;24(8):7662. doi: 10.3390/ijms24087662. Int J Mol Sci. 2023. PMID: 37108829 Free PMC article.
References
-
- Boonnak K., Suttitheptumrong A., Jotekratok U., Pattanakitsakul S. nga. Sphylogenetic analysis reveals genetic variations of densovirus isolated from field mosquitoes in Bangkok and surrounding regions. Southeast Asian J. Trop. Med. Publ. Health. 2015;46:207–214. - PubMed
-
- Breitbart M., Bonnain C., Malki K., Sawaya N.A. Phage puppet masters of the marine microbial realm. Nat. Microbiol. 2018;3:754–766. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources


