Histone variant H3.3 maintains adult haematopoietic stem cell homeostasis by enforcing chromatin adaptability
- PMID: 34961794
- PMCID: PMC9166935
- DOI: 10.1038/s41556-021-00795-7
Histone variant H3.3 maintains adult haematopoietic stem cell homeostasis by enforcing chromatin adaptability
Erratum in
-
Publisher Correction: Histone variant H3.3 maintains adult haematopoietic stem cell homeostasis by enforcing chromatin adaptability.Nat Cell Biol. 2022 Feb;24(2):279. doi: 10.1038/s41556-022-00851-w. Nat Cell Biol. 2022. PMID: 35058593 No abstract available.
Abstract
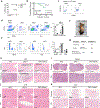
Histone variants and the associated post-translational modifications that govern the stemness of haematopoietic stem cells (HSCs) and differentiation thereof into progenitors (HSPCs) have not been well defined. H3.3 is a replication-independent H3 histone variant in mammalian systems that is enriched at both H3K4me3- and H3K27me3-marked bivalent genes as well as H3K9me3-marked endogenous retroviral repeats. Here we show that H3.3, but not its chaperone Hira, prevents premature HSC exhaustion and differentiation into granulocyte-macrophage progenitors. H3.3-null HSPCs display reduced expression of stemness and lineage-specific genes with a predominant gain of H3K27me3 marks at their promoter regions. Concomitantly, loss of H3.3 leads to a reduction of H3K9me3 marks at endogenous retroviral repeats, opening up binding sites for the interferon regulatory factor family of transcription factors, allowing the survival of rare, persisting H3.3-null HSCs. We propose a model whereby H3.3 maintains adult HSC stemness by safeguarding the delicate interplay between H3K27me3 and H3K9me3 marks, enforcing chromatin adaptability.
© 2021. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests
S.R. is the non-paid co-founder of Angiocrine Bioscience (San Diego). The other authors declare no competing interests.
Figures

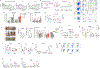













Comment in
-
H3.3 safeguards haematopoietic ERV-quilibrium.Nat Cell Biol. 2022 Jan;24(1):7-9. doi: 10.1038/s41556-021-00758-y. Nat Cell Biol. 2022. PMID: 34961795 No abstract available.
Similar articles
-
The histone H3.3 chaperone HIRA restrains erythroid-biased differentiation of adult hematopoietic stem cells.Stem Cell Reports. 2021 Aug 10;16(8):2014-2028. doi: 10.1016/j.stemcr.2021.06.009. Epub 2021 Jul 8. Stem Cell Reports. 2021. PMID: 34242617 Free PMC article.
-
HIRA, a DiGeorge Syndrome Candidate Gene, Confers Proper Chromatin Accessibility on HSCs and Supports All Stages of Hematopoiesis.Cell Rep. 2020 Feb 18;30(7):2136-2149.e4. doi: 10.1016/j.celrep.2020.01.062. Cell Rep. 2020. PMID: 32075733
-
H3.Y discriminates between HIRA and DAXX chaperone complexes and reveals unexpected insights into human DAXX-H3.3-H4 binding and deposition requirements.Nucleic Acids Res. 2017 Jun 2;45(10):5691-5706. doi: 10.1093/nar/gkx131. Nucleic Acids Res. 2017. PMID: 28334823 Free PMC article.
-
A Molecular Prospective for HIRA Complex Assembly and H3.3-Specific Histone Chaperone Function.J Mol Biol. 2017 Jun 30;429(13):1924-1933. doi: 10.1016/j.jmb.2016.11.010. Epub 2016 Nov 19. J Mol Biol. 2017. PMID: 27871933 Free PMC article. Review.
-
Bivalent Histone Modifications and Development.Curr Stem Cell Res Ther. 2018;13(2):83-90. doi: 10.2174/1574888X12666170123144743. Curr Stem Cell Res Ther. 2018. PMID: 28117006 Review.
Cited by
-
Roles of Histone H2B, H3 and H4 Variants in Cancer Development and Prognosis.Int J Mol Sci. 2024 Sep 7;25(17):9699. doi: 10.3390/ijms25179699. Int J Mol Sci. 2024. PMID: 39273649 Free PMC article. Review.
-
Trim47 prevents hematopoietic stem cell exhaustion during stress by regulating MAVS-mediated innate immune pathway.Nat Commun. 2024 Aug 8;15(1):6787. doi: 10.1038/s41467-024-51199-8. Nat Commun. 2024. PMID: 39117713 Free PMC article.
-
PTIP epigenetically regulates DNA damage-induced cell cycle arrest by upregulating PRDM1.Sci Rep. 2024 Aug 3;14(1):17987. doi: 10.1038/s41598-024-68295-w. Sci Rep. 2024. PMID: 39097652 Free PMC article.
-
Plasma cell differentiation is regulated by the expression of histone variant H3.3.Nat Commun. 2024 Jun 20;15(1):5004. doi: 10.1038/s41467-024-49375-x. Nat Commun. 2024. PMID: 38902223 Free PMC article.
-
KAT2A-mediated succinylation modification of notch1 promotes the proliferation and differentiation of dental pulp stem cells by activating notch pathway.BMC Oral Health. 2024 Mar 31;24(1):407. doi: 10.1186/s12903-024-03951-1. BMC Oral Health. 2024. PMID: 38556862 Free PMC article.
References
-
- Yu VWC et al. Epigenetic memory underlies cell-autonomous heterogeneous behavior of hematopoietic stem cells. Cell 167, 1310–1322 (2016). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases


