Tools used to assay genomic instability in cancers and cancer meiomitosis
- PMID: 34841477
- PMCID: PMC8891418
- DOI: 10.1007/s12079-021-00661-z
Tools used to assay genomic instability in cancers and cancer meiomitosis
Abstract
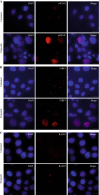
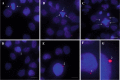
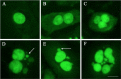
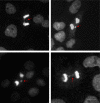
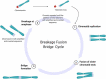
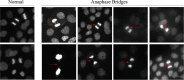
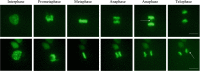
Genomic instability is a defining characteristic of cancer and the analysis of DNA damage at the chromosome level is a crucial part of the study of carcinogenesis and genotoxicity. Chromosomal instability (CIN), the most common level of genomic instability in cancers, is defined as the rate of loss or gain of chromosomes through successive divisions. As such, DNA in cancer cells is highly unstable. However, the underlying mechanisms remain elusive. There is a debate as to whether instability succeeds transformation, or if it is a by-product of cancer, and therefore, studying potential molecular and cellular contributors of genomic instability is of high importance. Recent work has suggested an important role for ectopic expression of meiosis genes in driving genomic instability via a process called meiomitosis. Improving understanding of these mechanisms can contribute to the development of targeted therapies that exploit DNA damage and repair mechanisms. Here, we discuss a workflow of novel and established techniques used to assess chromosomal instability as well as the nature of genomic instability such as double strand breaks, micronuclei, and chromatin bridges. For each technique, we discuss their advantages and limitations in a lab setting. Lastly, we provide detailed protocols for the discussed techniques.
Keywords: Chromosomal instability; Cytokinesis-block micronucleus assay; DNA damage; Genomic instability; H2B-GFP; Meiomitosis; Single-cell sequencing.
© 2021. The International CCN Society.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


Similar articles
-
Ectopically Expressed Meiosis-Specific Cancer Testis Antigen HORMAD1 Promotes Genomic Instability in Squamous Cell Carcinomas.Cells. 2023 Jun 14;12(12):1627. doi: 10.3390/cells12121627. Cells. 2023. PMID: 37371097 Free PMC article.
-
A study of meiomitosis and novel pathways of genomic instability in cutaneous T-cell lymphomas (CTCL).Oncotarget. 2018 Dec 28;9(102):37647-37661. doi: 10.18632/oncotarget.26479. eCollection 2018 Dec 28. Oncotarget. 2018. PMID: 30701021 Free PMC article.
-
Nuclear anomalies, chromosomal aberrations and proliferation rates in cultured lymphocytes of head and neck cancer patients.Asian Pac J Cancer Prev. 2014;15(3):1119-23. doi: 10.7314/apjcp.2014.15.3.1119. Asian Pac J Cancer Prev. 2014. PMID: 24606428
-
The ectopic expression of meiCT genes promotes meiomitosis and may facilitate carcinogenesis.Cell Cycle. 2020 Apr;19(8):837-854. doi: 10.1080/15384101.2020.1743902. Epub 2020 Mar 30. Cell Cycle. 2020. PMID: 32223693 Free PMC article. Review.
-
Cytokinesis-block micronucleus assay evolves into a "cytome" assay of chromosomal instability, mitotic dysfunction and cell death.Mutat Res. 2006 Aug 30;600(1-2):58-66. doi: 10.1016/j.mrfmmm.2006.05.028. Epub 2006 Jul 5. Mutat Res. 2006. PMID: 16822529 Review.
Cited by
-
Controversies regarding transplantation of mesenchymal stem cells.World J Transplant. 2024 Jun 18;14(2):90554. doi: 10.5500/wjt.v14.i2.90554. World J Transplant. 2024. PMID: 38947963 Free PMC article. Review.
-
The two sides of chromosomal instability: drivers and brakes in cancer.Signal Transduct Target Ther. 2024 Mar 29;9(1):75. doi: 10.1038/s41392-024-01767-7. Signal Transduct Target Ther. 2024. PMID: 38553459 Free PMC article. Review.
-
Plant Monoterpenes and Essential Oils as Potential Anti-Ageing Agents: Insights from Preclinical Data.Biomedicines. 2024 Feb 4;12(2):365. doi: 10.3390/biomedicines12020365. Biomedicines. 2024. PMID: 38397967 Free PMC article. Review.
-
The Ultraviolet Irradiation of Keratinocytes Induces Ectopic Expression of LINE-1 Retrotransposon Machinery and Leads to Cellular Senescence.Biomedicines. 2023 Nov 10;11(11):3017. doi: 10.3390/biomedicines11113017. Biomedicines. 2023. PMID: 38002016 Free PMC article.
-
Ectopically Expressed Meiosis-Specific Cancer Testis Antigen HORMAD1 Promotes Genomic Instability in Squamous Cell Carcinomas.Cells. 2023 Jun 14;12(12):1627. doi: 10.3390/cells12121627. Cells. 2023. PMID: 37371097 Free PMC article.
References
-
- Aaij R, Abellan Beteta C, Adeva B, Adinolfi M, Aidala CA, Ajaltouni Z, Akar S, Albicocco P, Albrecht J, Alessio F, et al. Measurement of antiproton production in p-He collisions at sqrt[s_{NN}]=110 GeV. Phys Rev Lett. 2018;121:222001. - PubMed
-
- Adelaide J, Finetti P, Bekhouche I, Repellini L, Geneix J, Sircoulomb F, Charafe-Jauffret E, Cervera N, Desplans J, Parzy D, et al. Integrated profiling of basal and luminal breast cancers. Cancer Res. 2007;67:11565–11575. - PubMed
-
- Albertson DG, Pinkel D. Genomic microarrays in human genetic disease and cancer. Hum Mol Genet. 2003;12(2):R145–R152. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous


