A Robust Panel Based on Mitochondrial Localized Proteins for Prognostic Prediction of Lung Adenocarcinoma
- PMID: 34539973
- PMCID: PMC8445726
- DOI: 10.1155/2021/7569168
A Robust Panel Based on Mitochondrial Localized Proteins for Prognostic Prediction of Lung Adenocarcinoma
Abstract
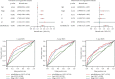
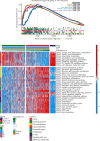
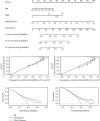
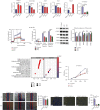
Due to high energy and material metabolism requirements, mitochondria are frequently active in tumor cells. Our study found that the high energy metabolism status is positively correlated with the poor prognosis of patients with lung adenocarcinoma. We constructed a scoring system (mitoRiskscore) based on the gene expression of specific mitochondrial localized proteins through univariate and LASSO cox regression. It has been shown that high mitoRiskscore was correlated with a shorter survival time after surgery in patients with lung adenocarcinoma. Compared with the typical TNM grading system, the mitoRiskscore gene panel had higher prediction accuracy. A vast number of external verification results ensured its universality. Additionally, the mitoRiskscore could evaluate the metabolic pattern and chemotherapy sensitivity of the tumor samples. Lung adenocarcinoma with higher mitoRiskscore was more active in glycolysis, and oxidative phosphorylation expression of proliferation-related pathway genes was also significantly upregulated. In contrast, patients with low mitoRiskscore had similar metabolic patterns to normal tissues. In order to improve the accuracy of prediction ability and promote clinical usage, we developed a nomogram that combined mitoRiskscore and clinical prognostic factors to predict the 3-year, 5-year, and 10-year survival rates of patients. We also performed in vitro experiments to verify the function of the key genes in the mitoRiskscore panel. In conclusion, the mitoRiskscore scoring system may assist clinicians to judge the postoperative survival rate and chemotherapy of patients with lung adenocarcinoma.
Copyright © 2021 Weifeng Chen et al.
Conflict of interest statement
All authors declare no financial competing interests.
Figures




Similar articles
-
Mitochondrial uncoupling protein 2 regulates the effects of paclitaxel on Stat3 activation and cellular survival in lung cancer cells.Carcinogenesis. 2012 Nov;33(11):2065-75. doi: 10.1093/carcin/bgs253. Epub 2012 Jul 30. Carcinogenesis. 2012. PMID: 22847181
-
Lactate dehydrogenase B is required for the growth of KRAS-dependent lung adenocarcinomas.Clin Cancer Res. 2013 Feb 15;19(4):773-84. doi: 10.1158/1078-0432.CCR-12-2638. Epub 2012 Dec 6. Clin Cancer Res. 2013. PMID: 23224736
-
Increased KIF15 Expression Predicts a Poor Prognosis in Patients with Lung Adenocarcinoma.Cell Physiol Biochem. 2018;51(1):1-10. doi: 10.1159/000495155. Epub 2018 Nov 15. Cell Physiol Biochem. 2018. PMID: 30439711
-
The mitochondrial voltage-dependent anion channel 1 in tumor cells.Biochim Biophys Acta. 2015 Oct;1848(10 Pt B):2547-75. doi: 10.1016/j.bbamem.2014.10.040. Epub 2014 Nov 4. Biochim Biophys Acta. 2015. PMID: 25448878 Review.
-
Prognosis and prognostic factors in adenocarcinoma of the lung.Dan Med Bull. 1992 Oct;39(5):453-63. Dan Med Bull. 1992. PMID: 1424819 Review. No abstract available.
Cited by
-
G6PD is a prognostic biomarker correlated with immune infiltrates in lung adenocarcinoma and pulmonary arterial hypertension.Aging (Albany NY). 2024 Jan 8;16(1):466-492. doi: 10.18632/aging.205381. Epub 2024 Jan 8. Aging (Albany NY). 2024. PMID: 38194707 Free PMC article.
References
-
- Goldstraw P., Chansky K., Crowley J., et al. The IASLC lung cancer staging project: proposals for revision of the TNM stage groupings in the forthcoming (eighth) edition of the TNM classification for lung cancer. Journal of Thoracic Oncology. 2016;11(1):39–51. doi: 10.1016/j.jtho.2015.09.009. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous


