Furin cleavage sites naturally occur in coronaviruses
- PMID: 33340798
- PMCID: PMC7836551
- DOI: 10.1016/j.scr.2020.102115
Furin cleavage sites naturally occur in coronaviruses
Abstract
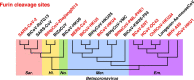
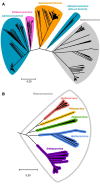
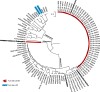
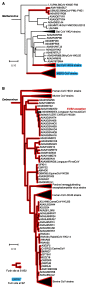
The spike protein is a focused target of COVID-19, a pandemic caused by SARS-CoV-2. A 12-nt insertion at S1/S2 in the spike coding sequence yields a furin cleavage site, which raised controversy views on origin of the virus. Here we analyzed the phylogenetic relationships of coronavirus spike proteins and mapped furin recognition motif on the tree. Furin cleavage sites occurred independently for multiple times in the evolution of the coronavirus family, supporting the natural occurring hypothesis of SARS-CoV-2.
Copyright © 2020 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

Similar articles
-
SARS-CoV-2 Spike Furin Cleavage Site and S2' Basic Residues Modulate the Entry Process in a Host Cell-Dependent Manner.J Virol. 2022 Jul 13;96(13):e0047422. doi: 10.1128/jvi.00474-22. Epub 2022 Jun 9. J Virol. 2022. PMID: 35678602 Free PMC article.
-
The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets.Nat Microbiol. 2021 Jul;6(7):899-909. doi: 10.1038/s41564-021-00908-w. Epub 2021 Apr 27. Nat Microbiol. 2021. PMID: 33907312
-
The sequence at Spike S1/S2 site enables cleavage by furin and phospho-regulation in SARS-CoV2 but not in SARS-CoV1 or MERS-CoV.Sci Rep. 2020 Oct 9;10(1):16944. doi: 10.1038/s41598-020-74101-0. Sci Rep. 2020. PMID: 33037310 Free PMC article.
-
Furin cleavage sites in the spike proteins of bat and rodent coronaviruses: Implications for virus evolution and zoonotic transfer from rodent species.One Health. 2021 Dec;13:100282. doi: 10.1016/j.onehlt.2021.100282. Epub 2021 Jun 22. One Health. 2021. PMID: 34179330 Free PMC article. Review.
-
Roles of the polybasic furin cleavage site of spike protein in SARS-CoV-2 replication, pathogenesis, and host immune responses and vaccination.J Med Virol. 2022 May;94(5):1815-1820. doi: 10.1002/jmv.27539. Epub 2021 Dec 31. J Med Virol. 2022. PMID: 34936124 Review.
Cited by
-
Will COVID-19 become mild, like a cold?Epidemiol Infect. 2024 Oct 7;152:e120. doi: 10.1017/S0950268824001110. Epidemiol Infect. 2024. PMID: 39370682 Free PMC article. Review.
-
Evaluation of Potential Furin Protease Inhibitory Properties of Pioglitazone, Rosiglitazone, and Pirfenidone: An In Silico Docking and Molecular Dynamics Simulation Approach.Curr Issues Mol Biol. 2024 Aug 8;46(8):8665-8684. doi: 10.3390/cimb46080511. Curr Issues Mol Biol. 2024. PMID: 39194728 Free PMC article.
-
Structural insights into Furin enzyme inhibition to block SARS-CoV-2 spike protein cleavage: an in-silico approach.3 Biotech. 2024 Sep;14(9):213. doi: 10.1007/s13205-024-04054-y. Epub 2024 Aug 25. 3 Biotech. 2024. PMID: 39193012
-
High-throughput screening identifies broad-spectrum Coronavirus entry inhibitors.iScience. 2024 May 17;27(6):110019. doi: 10.1016/j.isci.2024.110019. eCollection 2024 Jun 21. iScience. 2024. PMID: 38883823 Free PMC article.
-
Sequential glycosylations at the multibasic cleavage site of SARS-CoV-2 spike protein regulate viral activity.Nat Commun. 2024 May 16;15(1):4162. doi: 10.1038/s41467-024-48503-x. Nat Commun. 2024. PMID: 38755139 Free PMC article.
References
-
- Premkumar L., Segovia-Chumbez B., Jadi R., Martinez D.R., Raut R., Markmann A., Cornaby C., Bartelt L., Weiss S., Park Y., Edwards C.E., Weimer E., Scherer E.M., Rouphael N., Edupuganti S., Weiskopf D., Tse L.V., Hou Y.J., Margolis D., Sette A., Collins M.H., Schmitz J., Baric R.S., de Silva A.M. The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci. Immunol. 2020;5(48) - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous


