Evolutionary legacy of a forest plantation tree species (Pinus armandii): Implications for widespread afforestation
- PMID: 33294014
- PMCID: PMC7691453
- DOI: 10.1111/eva.13064
Evolutionary legacy of a forest plantation tree species (Pinus armandii): Implications for widespread afforestation
Abstract
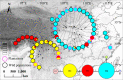
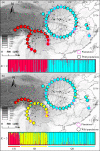
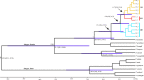
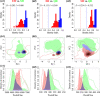
Many natural systems are subject to profound and persistent anthropogenic influence. Human-induced gene movement through afforestation and the selective transportation of genotypes might enhance the potential for intraspecific hybridization, which could lead to outbreeding depression. However, the evolutionary legacy of afforestation on the spatial genetic structure of forest tree species has barely been investigated. To do this properly, the effects of anthropogenic and natural processes must be examined simultaneously. A multidisciplinary approach, integrating phylogeography, population genetics, species distribution modeling, and niche divergence would permit evaluation of potential anthropogenic impacts, such as mass planting near-native material. Here, these approaches were applied to Pinus armandii, a Chinese endemic coniferous tree species, that has been mass planted across its native range. Population genetic analyses showed that natural populations of P. armandii comprised three lineages that diverged around the late Miocene, during a period of massive uplifts of the Hengduan Mountains, and intensification of Asian Summer Monsoon. Only limited gene flow was detected between lineages, indicating that each largely maintained is genetic integrity. Moreover, most or all planted populations were found to have been sourced within the same region, minimizing disruption of large-scale spatial genetic structure within P. armandii. This might be because each of the three lineages had a distinct climatic niche, according to ecological niche modeling and niche divergence tests. The current study provides empirical genetic and ecological evidence for the site-species matching principle in forestry and will be useful to manage restoration efforts by identifying suitable areas and climates for introducing and planting new forests. Our results also highlight the urgent need to evaluate the genetic impacts of large-scale afforestation in other native tree species.
Keywords: Pinus armandii; afforestation; forest plantations; niche divergence; spatial genetic structure.
© 2020 The Authors. Evolutionary Applications published by John Wiley & Sons Ltd.
Figures

Similar articles
-
[Resources utilization of main tree populations in Pinus armandii mixed forest after controlled burning and aerial-sowing afforestation].Ying Yong Sheng Tai Xue Bao. 2005 Apr;16(4):600-4. Ying Yong Sheng Tai Xue Bao. 2005. PMID: 16011151 Chinese.
-
Cryptic speciation in the Chinese white pine (Pinus armandii): Implications for the high species diversity of conifers in the Hengduan Mountains, a global biodiversity hotspot.Mol Phylogenet Evol. 2019 Sep;138:114-125. doi: 10.1016/j.ympev.2019.05.015. Epub 2019 May 18. Mol Phylogenet Evol. 2019. PMID: 31112783
-
Phylogeography of Pinus armandii and its relatives: heterogeneous contributions of geography and climate changes to the genetic differentiation and diversification of Chinese white pines.PLoS One. 2014 Jan 21;9(1):e85920. doi: 10.1371/journal.pone.0085920. eCollection 2014. PLoS One. 2014. PMID: 24465789 Free PMC article.
-
Transitional forestry in New Zealand: re-evaluating the design and management of forest systems through the lens of forest purpose.Biol Rev Camb Philos Soc. 2023 Aug;98(4):1003-1015. doi: 10.1111/brv.12941. Epub 2023 Feb 19. Biol Rev Camb Philos Soc. 2023. PMID: 36808687 Review.
-
Ecological impacts of non-native tree species plantations are broad and heterogeneous: a review of Brazilian research.An Acad Bras Cienc. 2016;88(3 Suppl):1675-1688. doi: 10.1590/0001-3765201620150575. Epub 2016 Oct 10. An Acad Bras Cienc. 2016. PMID: 27737335 Review. Portuguese.
Cited by
-
Guiding seed movement: environmental heterogeneity drives genetic differentiation in Plathymenia reticulata, providing insights for restoration.AoB Plants. 2024 May 29;16(3):plae032. doi: 10.1093/aobpla/plae032. eCollection 2024 Jun. AoB Plants. 2024. PMID: 38883565 Free PMC article.
-
Population differentiation and dynamics of five pioneer species of Gaultheria from the secondary forests in subtropical China.BMC Plant Biol. 2024 Jun 8;24(1):516. doi: 10.1186/s12870-024-05189-z. BMC Plant Biol. 2024. PMID: 38851686 Free PMC article.
-
The complete chloroplast genome sequence of Pinus bhutanica (Pinaceae) and its phylogenetic implications.Mitochondrial DNA B Resour. 2024 Jan 26;9(1):182-185. doi: 10.1080/23802359.2024.2305710. eCollection 2024. Mitochondrial DNA B Resour. 2024. PMID: 38288249 Free PMC article.
-
Genomic insights into differentiation and adaptation of Amorphophallus yunnanensis in the mountainous region of Southwest China.Ecol Evol. 2024 Jan 23;14(1):e10861. doi: 10.1002/ece3.10861. eCollection 2024 Jan. Ecol Evol. 2024. PMID: 38264337 Free PMC article.
-
Biomod2 modeling for predicting the potential ecological distribution of three Fritillaria species under climate change.Sci Rep. 2023 Nov 1;13(1):18801. doi: 10.1038/s41598-023-45887-6. Sci Rep. 2023. PMID: 37914761 Free PMC article.
References
-
- An, Z. S. , Sun, Y. B. , Chang, H. , Zhang, P. Z. , Liu, X. D. , Cai, Y. J. , … Zhao, J. (2014). Late Cenozoic climate change in monsoon‐arid Asia and global changes In An Z. S. (Ed.), Late Cenozoic Climate Change in Asia (pp. 491–581). Berlin: Springer.
-
- Antonelli, A. , Kissling, W. D. , Flantua, S. G. A. , Bermúdez, M. A. , Mulch, A. , Muellner‐Riehl, A. N. , … Hoorn, C. (2018). Geological and climatic influences on mountain biodiversity. Nature Geoscience, 11, 718–725. 10.1038/s41561-018-0236-z - DOI
-
- Avise, J. C. (2000). Phylogeography: The history and formation of species. Cambridge, UK: Harvard University Press.
Associated data
LinkOut - more resources
Full Text Sources


