The Chd1 chromatin remodeler forms long-lived complexes with nucleosomes in the presence of ADP·BeF3- and transition state analogs
- PMID: 31636125
- PMCID: PMC6885614
- DOI: 10.1074/jbc.RA119.009782
The Chd1 chromatin remodeler forms long-lived complexes with nucleosomes in the presence of ADP·BeF3- and transition state analogs
Abstract
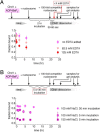
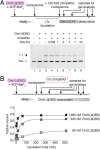
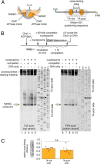
Chromatin remodelers use helicase-like ATPase domains to reorganize histone-DNA contacts within the nucleosome. Like other remodelers, the chromodomain helicase DNA-binding protein 1 (Chd1) remodeler repositions nucleosomes by altering DNA topology at its internal binding site on the nucleosome, coupling different degrees of DNA twist and DNA movement to distinct nucleotide-bound states of the ATPase motor. In this work, we used a competition assay to study how variations in the bound nucleotide, Chd1, and the nucleosome substrate affect stability of Chd1-nucleosome complexes. We found that Chd1-nucleosome complexes formed in nucleotide-free or ADP conditions were relatively unstable and dissociated within 30 s, whereas those with the nonhydrolyzable ATP analog AMP-PNP had a mean lifetime of 4.8 ± 0.7 min. Chd1-nucleosome complexes were remarkably stable with ADP·BeF3- and the transition state analogs ADP·AlFX and ADP·MgFX, being resistant to competitor nucleosome over a 24-h period. For the tight ADP·BeF3--stabilized complex, Mg2+ was a critical component that did not freely exchange, and formation of these long-lived complexes had a slow, concentration-dependent step. The ADP·BeF3--stabilized complex did not require the Chd1 DNA-binding domain nor the histone H4 tail and appeared relatively insensitive to sequence differences on either side of the Widom 601 sequence. Interestingly, the complex remained stable in ADP·BeF3- even when nucleosomes contained single-stranded gaps that disrupted most DNA contacts with the guide strand. This finding suggests that binding via the tracking strand alone is sufficient for stabilizing the complex in a hydrolysis-competent state.
Keywords: ATPase; DNA binding protein; chromatin remodeling; chromodomain helicase DNA-binding protein 1 (Chd1); motor protein; nucleoside/nucleotide analog; nucleosome; superfamily 2 (SF2) ATPase; transition state analog; twist defect.
© 2019 Ren et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



Similar articles
-
The ATPase motor of the Chd1 chromatin remodeler stimulates DNA unwrapping from the nucleosome.Nucleic Acids Res. 2018 Jun 1;46(10):4978-4990. doi: 10.1093/nar/gky206. Nucleic Acids Res. 2018. PMID: 29850894 Free PMC article.
-
Nucleosome recognition and DNA distortion by the Chd1 remodeler in a nucleotide-free state.Nat Struct Mol Biol. 2022 Feb;29(2):121-129. doi: 10.1038/s41594-021-00719-x. Epub 2022 Feb 16. Nat Struct Mol Biol. 2022. PMID: 35173352 Free PMC article.
-
A twist defect mechanism for ATP-dependent translocation of nucleosomal DNA.Elife. 2018 May 29;7:e34100. doi: 10.7554/eLife.34100. Elife. 2018. PMID: 29809147 Free PMC article.
-
Genome-Wide Analysis of Nucleosome Positions, Occupancy, and Accessibility in Yeast: Nucleosome Mapping, High-Resolution Histone ChIP, and NCAM.Curr Protoc Mol Biol. 2014 Oct 1;108:21.28.1-21.28.16. doi: 10.1002/0471142727.mb2128s108. Curr Protoc Mol Biol. 2014. PMID: 25271716 Free PMC article. Review.
-
Yeast HMO1: Linker Histone Reinvented.Microbiol Mol Biol Rev. 2016 Nov 30;81(1):e00037-16. doi: 10.1128/MMBR.00037-16. Print 2017 Mar. Microbiol Mol Biol Rev. 2016. PMID: 27903656 Free PMC article. Review.
Cited by
-
H2A.Z deposition by SWR1C involves multiple ATP-dependent steps.Nat Commun. 2022 Nov 17;13(1):7052. doi: 10.1038/s41467-022-34861-x. Nat Commun. 2022. PMID: 36396651 Free PMC article.
-
SMARCAD1 is an ATP-dependent histone octamer exchange factor with de novo nucleosome assembly activity.Sci Adv. 2021 Oct 15;7(42):eabk2380. doi: 10.1126/sciadv.abk2380. Epub 2021 Oct 15. Sci Adv. 2021. PMID: 34652950 Free PMC article.
-
Autoinhibitory elements of the Chd1 remodeler block initiation of twist defects by destabilizing the ATPase motor on the nucleosome.Proc Natl Acad Sci U S A. 2021 Jan 26;118(4):e2014498118. doi: 10.1073/pnas.2014498118. Proc Natl Acad Sci U S A. 2021. PMID: 33468676 Free PMC article.
-
Architecture of the herpesvirus genome-packaging complex and implications for DNA translocation.Protein Cell. 2020 May;11(5):339-351. doi: 10.1007/s13238-020-00710-0. Epub 2020 Apr 23. Protein Cell. 2020. PMID: 32328903 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous


