Endemic infection can shape exposure to novel pathogens: Pathogen co-occurrence networks in the Serengeti lions
- PMID: 30861289
- PMCID: PMC7163671
- DOI: 10.1111/ele.13250
Endemic infection can shape exposure to novel pathogens: Pathogen co-occurrence networks in the Serengeti lions
Abstract
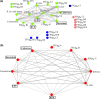
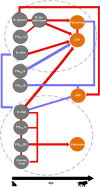
Pathogens are embedded in a complex network of microparasites that can collectively or individually alter disease dynamics and outcomes. Endemic pathogens that infect an individual in the first years of life, for example, can either facilitate or compete with subsequent pathogens thereby exacerbating or ameliorating morbidity and mortality. Pathogen associations are ubiquitous but poorly understood, particularly in wild populations. We report here on 10 years of serological and molecular data in African lions, leveraging comprehensive demographic and behavioural data to test if endemic pathogens shape subsequent infection by epidemic pathogens. We combine network and community ecology approaches to assess broad network structure and characterise associations between pathogens across spatial and temporal scales. We found significant non-random structure in the lion-pathogen co-occurrence network and identified both positive and negative associations between endemic and epidemic pathogens. Our results provide novel insights on the complex associations underlying pathogen co-occurrence networks.
Keywords: Babesia; calicivirus; canine distemper virus; co-infection; community assembly; coronavirus; feline immunodeficiency virus; parvovirus.
© 2019 John Wiley & Sons Ltd/CNRS.
Figures

Similar articles
-
SURVEILLANCE FOR VIRAL AND PARASITIC PATHOGENS IN A VULNERABLE AFRICAN LION (PANTHERA LEO) POPULATION IN THE NORTHERN TULI GAME RESERVE, BOTSWANA.J Wildl Dis. 2017 Jan;53(1):54-61. doi: 10.7589/2015-09-248. Epub 2016 Sep 26. J Wildl Dis. 2017. PMID: 27669009
-
Linking social and spatial networks to viral community phylogenetics reveals subtype-specific transmission dynamics in African lions.J Anim Ecol. 2017 Oct;86(6):1469-1482. doi: 10.1111/1365-2656.12751. Epub 2017 Oct 10. J Anim Ecol. 2017. PMID: 28884827
-
Climate extremes promote fatal co-infections during canine distemper epidemics in African lions.PLoS One. 2008 Jun 25;3(6):e2545. doi: 10.1371/journal.pone.0002545. PLoS One. 2008. PMID: 18575601 Free PMC article.
-
Mycobacterium bovis infection in the lion (Panthera leo): Current knowledge, conundrums and research challenges.Vet Microbiol. 2015 Jun 12;177(3-4):252-60. doi: 10.1016/j.vetmic.2015.03.028. Epub 2015 Apr 8. Vet Microbiol. 2015. PMID: 25891424 Review.
-
The role of social structure and dynamics in the maintenance of endemic disease.Behav Ecol Sociobiol. 2021;75(8):122. doi: 10.1007/s00265-021-03055-8. Epub 2021 Aug 18. Behav Ecol Sociobiol. 2021. PMID: 34421183 Free PMC article. Review.
Cited by
-
A systematic review and guide for using multi-response statistical models in co-infection research.R Soc Open Sci. 2024 Oct 4;11(10):231589. doi: 10.1098/rsos.231589. eCollection 2024 Oct. R Soc Open Sci. 2024. PMID: 39371046 Free PMC article.
-
Bacterial Community Characteristics Shaped by Artificial Environmental PM2.5 Control in Intensive Broiler Houses.Int J Environ Res Public Health. 2022 Dec 30;20(1):723. doi: 10.3390/ijerph20010723. Int J Environ Res Public Health. 2022. PMID: 36613044 Free PMC article.
-
Coinfecting parasites can modify fluctuating selection dynamics in host-parasite coevolution.Ecol Evol. 2020 Aug 30;10(18):9600-9612. doi: 10.1002/ece3.6373. eCollection 2020 Sep. Ecol Evol. 2020. PMID: 33005333 Free PMC article.
-
Parasite associations predict infection risk: incorporating co-infections in predictive models for neglected tropical diseases.Parasit Vectors. 2020 Mar 16;13(1):138. doi: 10.1186/s13071-020-04016-2. Parasit Vectors. 2020. PMID: 32178706 Free PMC article.
References
-
- Aivelo, T. & Norberg, A. (2018). Parasite‐microbiota interactions potentially affect intestinal communities in wild mammals. J. Anim. Ecol., 87, 438–447. - PubMed
-
- Araújo, M.B. & Rozenfeld, A. (2014). The geographic scaling of biotic interactions. Ecography (Cop.), 37, 406–415.
-
- Benesh, D.P. & Kalbe, M. (2016). Experimental parasite community ecology: intraspecific variation in a large tapeworm affects community assembly. J. Anim. Ecol., 85, 1004–1013. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources


