Chromatin remodelers couple inchworm motion with twist-defect formation to slide nucleosomal DNA
- PMID: 30395604
- PMCID: PMC6237416
- DOI: 10.1371/journal.pcbi.1006512
Chromatin remodelers couple inchworm motion with twist-defect formation to slide nucleosomal DNA
Abstract
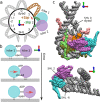
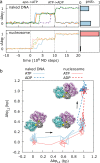
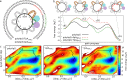
ATP-dependent chromatin remodelers are molecular machines that control genome organization by repositioning, ejecting, or editing nucleosomes, activities that confer them essential regulatory roles on gene expression and DNA replication. Here, we investigate the molecular mechanism of active nucleosome sliding by means of molecular dynamics simulations of the Snf2 remodeler translocase in complex with a nucleosome. During its inchworm motion driven by ATP consumption, the translocase overwrites the original nucleosome energy landscape via steric and electrostatic interactions to induce sliding of nucleosomal DNA unidirectionally. The sliding is initiated at the remodeler binding location via the generation of a pair of twist defects, which then spontaneously propagate to complete sliding throughout the entire nucleosome. We also reveal how remodeler mutations and DNA sequence control active nucleosome repositioning, explaining several past experimental observations. These results offer a detailed mechanistic picture of remodeling important for the complete understanding of these key biological processes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
A twist defect mechanism for ATP-dependent translocation of nucleosomal DNA.Elife. 2018 May 29;7:e34100. doi: 10.7554/eLife.34100. Elife. 2018. PMID: 29809147 Free PMC article.
-
Sequence-targeted nucleosome sliding in vivo by a hybrid Chd1 chromatin remodeler.Genome Res. 2016 May;26(5):693-704. doi: 10.1101/gr.199919.115. Epub 2016 Mar 18. Genome Res. 2016. PMID: 26993344 Free PMC article.
-
Mechanisms of ATP-dependent nucleosome sliding.Curr Opin Struct Biol. 2010 Feb;20(1):73-81. doi: 10.1016/j.sbi.2009.12.002. Epub 2010 Jan 8. Curr Opin Struct Biol. 2010. PMID: 20060707 Free PMC article. Review.
-
Distortion of histone octamer core promotes nucleosome mobilization by a chromatin remodeler.Science. 2017 Jan 20;355(6322):eaaa3761. doi: 10.1126/science.aaa3761. Science. 2017. PMID: 28104838 Free PMC article.
-
Targeting chromatin remodelers: signals and search mechanisms.Biochim Biophys Acta. 2011 Sep;1809(9):497-508. doi: 10.1016/j.bbagrm.2011.06.005. Epub 2011 Jun 16. Biochim Biophys Acta. 2011. PMID: 21704204 Review.
Cited by
-
Local volume concentration, packing domains, and scaling properties of chromatin.Elife. 2024 Sep 27;13:RP97604. doi: 10.7554/eLife.97604. Elife. 2024. PMID: 39331520 Free PMC article.
-
Integrative Modeling of 3D Genome Organization by Bayesian Molecular Dynamics Simulations with Hi-C Metainference.Methods Mol Biol. 2025;2856:309-324. doi: 10.1007/978-1-0716-4136-1_19. Methods Mol Biol. 2025. PMID: 39283461
-
Chromatin remodelers: a concise introduction for biophysicists.Biophys Rev. 2024 Jun 6;16(3):357-363. doi: 10.1007/s12551-024-01199-1. eCollection 2024 Jun. Biophys Rev. 2024. PMID: 39099840 Review.
-
Local Volume Concentration, Packing Domains and Scaling Properties of Chromatin.ArXiv [Preprint]. 2024 Jun 13:arXiv:2310.02257v3. ArXiv. 2024. Update in: Elife. 2024 Sep 27;13:RP97604. doi: 10.7554/eLife.97604. PMID: 38495560 Free PMC article. Updated. Preprint.
-
Switching Go̅-Martini for Investigating Protein Conformational Transitions and Associated Protein-Lipid Interactions.J Chem Theory Comput. 2024 Mar 26;20(6):2618-2629. doi: 10.1021/acs.jctc.3c01222. Epub 2024 Mar 6. J Chem Theory Comput. 2024. PMID: 38447049 Free PMC article.
References
-
- Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. Nature Publishing Group; 2003;423: 145–150. 10.1038/nature01595 - DOI - PubMed
-
- Lai WKM, Pugh BF. Understanding nucleosome dynamics and their links to gene expression and DNA replication. Nat Rev Mol Cell Biol. Nature Publishing Group; 2017;18: 548–562. 10.1038/nrm.2017.47 - DOI - PMC - PubMed
-
- Field Y, Kaplan N, Fondufe-Mittendorf Y, Moore IK, Sharon E, Lubling Y, et al. Distinct Modes of Regulation by Chromatin Encoded through Nucleosome Positioning Signals. Ohler U, editor. PLoS Comput Biol. Public Library of Science; 2008;4: e1000216 10.1371/journal.pcbi.1000216 - DOI - PMC - PubMed
-
- Struhl K, Segal E. Determinants of nucleosome positioning. Nat Struct Mol Biol. 2013;20: 267–273. 10.1038/nsmb.2506 - DOI - PMC - PubMed
-
- Teif VB, Vainshtein Y, Caudron-Herger M, Mallm J-P, Marth C, Höfer T, et al. Genome-wide nucleosome positioning during embryonic stem cell development. Nat Struct Mol Biol. Nature Publishing Group; 2012;19: 1185–1192. 10.1038/nsmb.2419 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous


