Carriers of mitochondrial DNA macrohaplogroup L3 basal lineages migrated back to Africa from Asia around 70,000 years ago
- PMID: 29921229
- PMCID: PMC6009813
- DOI: 10.1186/s12862-018-1211-4
Carriers of mitochondrial DNA macrohaplogroup L3 basal lineages migrated back to Africa from Asia around 70,000 years ago
Abstract
Background: The main unequivocal conclusion after three decades of phylogeographic mtDNA studies is the African origin of all extant modern humans. In addition, a southern coastal route has been argued for to explain the Eurasian colonization of these African pioneers. Based on the age of macrohaplogroup L3, from which all maternal Eurasian and the majority of African lineages originated, the out-of-Africa event has been dated around 60-70 kya. On the opposite side, we have proposed a northern route through Central Asia across the Levant for that expansion and, consistent with the fossil record, we have dated it around 125 kya. To help bridge differences between the molecular and fossil record ages, in this article we assess the possibility that mtDNA macrohaplogroup L3 matured in Eurasia and returned to Africa as basal L3 lineages around 70 kya.
Results: The coalescence ages of all Eurasian (M,N) and African (L3 ) lineages, both around 71 kya, are not significantly different. The oldest M and N Eurasian clades are found in southeastern Asia instead near of Africa as expected by the southern route hypothesis. The split of the Y-chromosome composite DE haplogroup is very similar to the age of mtDNA L3. An Eurasian origin and back migration to Africa has been proposed for the African Y-chromosome haplogroup E. Inside Africa, frequency distributions of maternal L3 and paternal E lineages are positively correlated. This correlation is not fully explained by geographic or ethnic affinities. This correlation rather seems to be the result of a joint and global replacement of the old autochthonous male and female African lineages by the new Eurasian incomers.
Conclusions: These results are congruent with a model proposing an out-of-Africa migration into Asia, following a northern route, of early anatomically modern humans carrying pre-L3 mtDNA lineages around 125 kya, subsequent diversification of pre-L3 into the basal lineages of L3, a return to Africa of Eurasian fully modern humans around 70 kya carrying the basal L3 lineages and the subsequent diversification of Eurasian-remaining L3 lineages into the M and N lineages in the outside-of-Africa context, and a second Eurasian global expansion by 60 kya, most probably, out of southeast Asia. Climatic conditions and the presence of Neanderthals and other hominins might have played significant roles in these human movements. Moreover, recent studies based on ancient DNA and whole-genome sequencing are also compatible with this hypothesis.
Keywords: Haplogroup E; Haplogroup L3; Human evolution; Mitochondrial DNA; Out-of-Africa; Y-chromosome.
Conflict of interest statement
Authors’ information
VMC is actually retired.
Ethics approval and consent to participate
The procedure of human population sampling adhered to the tenets of the Declaration of Helsinki. Written consent was recorded from all participants prior to taking part in the study. The study underwent formal review and was approved by the College of Medicine Ethical Committee of the King Saud University (proposal N°09-659), and by the Ethics Committee for Human Research at the University of La Laguna (proposal NR157).
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
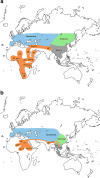
Similar articles
-
Carriers of human mitochondrial DNA macrohaplogroup M colonized India from southeastern Asia.BMC Evol Biol. 2016 Nov 10;16(1):246. doi: 10.1186/s12862-016-0816-8. BMC Evol Biol. 2016. PMID: 27832758 Free PMC article.
-
Carriers of mitochondrial DNA macrohaplogroup R colonized Eurasia and Australasia from a southeast Asia core area.BMC Evol Biol. 2017 May 23;17(1):115. doi: 10.1186/s12862-017-0964-5. BMC Evol Biol. 2017. PMID: 28535779 Free PMC article.
-
Carriers of Mitochondrial DNA Macrohaplogroup N Lineages Reached Australia around 50,000 Years Ago following a Northern Asian Route.PLoS One. 2015 Jun 8;10(6):e0129839. doi: 10.1371/journal.pone.0129839. eCollection 2015. PLoS One. 2015. PMID: 26053380 Free PMC article.
-
African human mtDNA phylogeography at-a-glance.J Anthropol Sci. 2011;89:25-58. doi: 10.4436/jass.89006. Epub 2011 Mar 15. J Anthropol Sci. 2011. PMID: 21368343 Review.
-
The reversal of human phylogeny: Homo left Africa as erectus, came back as sapiens sapiens.Hereditas. 2020 Dec 19;157(1):51. doi: 10.1186/s41065-020-00163-9. Hereditas. 2020. PMID: 33341120 Free PMC article. Review.
Cited by
-
A Customized Human Mitochondrial DNA Database (hMITO DB v1.0) for Rapid Sequence Analysis, Haplotyping and Geo-Mapping.Int J Mol Sci. 2023 Aug 31;24(17):13505. doi: 10.3390/ijms241713505. Int J Mol Sci. 2023. PMID: 37686313 Free PMC article.
-
North and East African mitochondrial genetic variation needs further characterization towards precision medicine.J Adv Res. 2023 Dec;54:59-76. doi: 10.1016/j.jare.2023.01.021. Epub 2023 Feb 2. J Adv Res. 2023. PMID: 36736695 Free PMC article.
-
Evaluation of D-loop hypervariable region I variations, haplogroups and copy number of mitochondrial DNA in Bangladeshi population with type 2 diabetes.Heliyon. 2021 Jul 15;7(7):e07573. doi: 10.1016/j.heliyon.2021.e07573. eCollection 2021 Jul. Heliyon. 2021. PMID: 34377852 Free PMC article.
-
Delineation of Mitochondrial DNA Variants From Exome Sequencing Data and Association of Haplogroups With Obesity in Kuwait.Front Genet. 2021 Feb 11;12:626260. doi: 10.3389/fgene.2021.626260. eCollection 2021. Front Genet. 2021. PMID: 33659027 Free PMC article.
-
Human molecular evolutionary rate, time dependency and transient polymorphism effects viewed through ancient and modern mitochondrial DNA genomes.Sci Rep. 2021 Mar 3;11(1):5036. doi: 10.1038/s41598-021-84583-1. Sci Rep. 2021. PMID: 33658608 Free PMC article.
References
-
- Cann RL, Stoneking M, Wilson AC. Mitochondrial DNA and human evolution. Nature. 325:31–6. - PubMed
-
- Herrnstadt C, Elson JL, Fahy E, Preston G, Turnbull DM, Anderson C, Ghosh SS, Olefsky JM, Beal MF, Davis RE, Howell N. Reduced-median-network analysis of complete mitochondrial DNA coding-region sequences for the major African, Asian, and European haplogroups. Am J Hum Genet. 2002;70:1152–1171. doi: 10.1086/339933. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources


