Repair of 8-oxoG:A mismatches by the MUTYH glycosylase: Mechanism, metals and medicine
- PMID: 28087410
- PMCID: PMC5457711
- DOI: 10.1016/j.freeradbiomed.2017.01.008
Repair of 8-oxoG:A mismatches by the MUTYH glycosylase: Mechanism, metals and medicine
Abstract
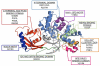
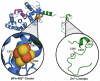
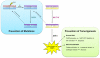
Reactive oxygen and nitrogen species (RONS) may infringe on the passing of pristine genetic information by inducing DNA inter- and intra-strand crosslinks, protein-DNA crosslinks, and chemical alterations to the sugar or base moieties of DNA. 8-Oxo-7,8-dihydroguanine (8-oxoG) is one of the most prevalent DNA lesions formed by RONS and is repaired through the base excision repair (BER) pathway involving the DNA repair glycosylases OGG1 and MUTYH in eukaryotes. MUTYH removes adenine (A) from 8-oxoG:A mispairs, thus mitigating the potential of G:C to T:A transversion mutations from occurring in the genome. The paramount role of MUTYH in guarding the genome is well established in the etiology of a colorectal cancer predisposition syndrome involving variants of MUTYH, referred to as MUTYH-associated polyposis (MAP). In this review, we highlight recent advances in understanding how MUTYH structure and related function participate in the manifestation of human disease such as MAP. Here we focus on the importance of MUTYH's metal cofactor sites, including a recently discovered "Zinc linchpin" motif, as well as updates to the catalytic mechanism. Finally, we touch on the insight gleaned from studies with MAP-associated MUTYH variants and recent advances in understanding the multifaceted roles of MUTYH in the cell, both in the prevention of mutagenesis and tumorigenesis.
Keywords: 8-oxoguanine; Base excision repair; Fe-S clusters; Glycosylase; MUTYH; MUTYH-associated polyposis; MutY.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures



Similar articles
-
Repair of 8-oxo-7,8-dihydroguanine in prokaryotic and eukaryotic cells: Properties and biological roles of the Fpg and OGG1 DNA N-glycosylases.Free Radic Biol Med. 2017 Jun;107:179-201. doi: 10.1016/j.freeradbiomed.2016.11.042. Epub 2016 Nov 27. Free Radic Biol Med. 2017. PMID: 27903453 Review.
-
Functional analysis of MUTYH mutated proteins associated with familial adenomatous polyposis.DNA Repair (Amst). 2010 Jun 4;9(6):700-7. doi: 10.1016/j.dnarep.2010.03.008. Epub 2010 Apr 24. DNA Repair (Amst). 2010. PMID: 20418187
-
Single molecule glycosylase studies with engineered 8-oxoguanine DNA damage sites show functional defects of a MUTYH polyposis variant.Nucleic Acids Res. 2019 Apr 8;47(6):3058-3071. doi: 10.1093/nar/gkz045. Nucleic Acids Res. 2019. PMID: 30698731 Free PMC article.
-
Distinct functional consequences of MUTYH variants associated with colorectal cancer: Damaged DNA affinity, glycosylase activity and interaction with PCNA and Hus1.DNA Repair (Amst). 2015 Oct;34:39-51. doi: 10.1016/j.dnarep.2015.08.001. Epub 2015 Aug 12. DNA Repair (Amst). 2015. PMID: 26377631 Free PMC article.
-
MutYH (MYH) and colorectal cancer.Biochem Soc Trans. 2005 Aug;33(Pt 4):679-83. doi: 10.1042/BST0330679. Biochem Soc Trans. 2005. PMID: 16042573 Review.
Cited by
-
Understanding Active Photoprotection: DNA-Repair Enzymes and Antioxidants.Life (Basel). 2024 Jun 28;14(7):822. doi: 10.3390/life14070822. Life (Basel). 2024. PMID: 39063576 Free PMC article. Review.
-
The interplay of DNA damage and repair, gene expression, and mutagenesis in mammalian cells during oxidative stress.Carcinogenesis. 2024 Nov 22;45(11):868-879. doi: 10.1093/carcin/bgae046. Carcinogenesis. 2024. PMID: 39023127
-
FSHing for DNA Damage: Key Features of MutY Detection of 8-Oxoguanine:Adenine Mismatches.Acc Chem Res. 2024 Apr 2;57(7):1019-1031. doi: 10.1021/acs.accounts.3c00759. Epub 2024 Mar 12. Acc Chem Res. 2024. PMID: 38471078 Free PMC article. Review.
-
Cellular Repair of Synthetic Analogs of Oxidative DNA Damage Reveals a Key Structure-Activity Relationship of the Cancer-Associated MUTYH DNA Repair Glycosylase.ACS Cent Sci. 2024 Jan 26;10(2):291-301. doi: 10.1021/acscentsci.3c00784. eCollection 2024 Feb 28. ACS Cent Sci. 2024. PMID: 38435525 Free PMC article.
-
A maternal germline mutator phenotype in a family affected by heritable colorectal cancer.medRxiv [Preprint]. 2024 Oct 4:2023.12.08.23299304. doi: 10.1101/2023.12.08.23299304. medRxiv. 2024. Update in: Genetics. 2024 Oct 15:iyae166. doi: 10.1093/genetics/iyae166 PMID: 38196581 Free PMC article. Updated. Preprint.
References
-
- Su SS, Lahue RS, Au KG, Modrich P. Mispair specificity of methyl-directed DNA mismatch correction in vitro. The Journal of biological chemistry. 1988;263(14):6829–35. - PubMed
-
- David SS, Wiliams SD. Chemistry of glycosylases and endonucleases involved in base-excision repair. Chemical reviews. 1998;98(3):1221–1261. - PubMed
-
- Lindahl T. Uracil-DNA glycosylase from Escherichia coli. Methods Enzymol. 1980;65(1):284–90. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials


