The Cultivable Surface Microbiota of the Brown Alga Ascophyllum nodosum is Enriched in Macroalgal-Polysaccharide-Degrading Bacteria
- PMID: 26734000
- PMCID: PMC4690005
- DOI: 10.3389/fmicb.2015.01487
The Cultivable Surface Microbiota of the Brown Alga Ascophyllum nodosum is Enriched in Macroalgal-Polysaccharide-Degrading Bacteria
Abstract
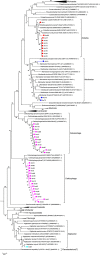
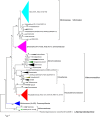
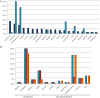
Bacteria degrading algal polysaccharides are key players in the global carbon cycle and in algal biomass recycling. Yet the water column, which has been studied largely by metagenomic approaches, is poor in such bacteria and their algal-polysaccharide-degrading enzymes. Even more surprisingly, the few published studies on seaweed-associated microbiomes have revealed low abundances of such bacteria and their specific enzymes. However, as macroalgal cell-wall polysaccharides do not accumulate in nature, these bacteria and their unique polysaccharidases must not be that uncommon. We, therefore, looked at the polysaccharide-degrading activity of the cultivable bacterial subpopulation associated with Ascophyllum nodosum. From A. nodosum triplicates, 324 bacteria were isolated and taxonomically identified. Out of these isolates, 78 (~25%) were found to act on at least one tested algal polysaccharide (agar, ι- or κ-carrageenan, or alginate). The isolates "active" on algal-polysaccharides belong to 11 genera: Cellulophaga, Maribacter, Algibacter, and Zobellia in the class Flavobacteriia (41) and Pseudoalteromonas, Vibrio, Cobetia, Shewanella, Colwellia, Marinomonas, and Paraglaceciola in the class Gammaproteobacteria (37). A major part represents likely novel species. Different proportions of bacterial phyla and classes were observed between the isolated cultivable subpopulation and the total microbial community previously identified on other brown algae. Here, Bacteroidetes and Gammaproteobacteria were found to be the most abundant and some phyla (as Planctomycetes and Cyanobacteria) frequently encountered on brown algae weren't identified. At a lower taxonomic level, twelve genera, well-known to be associated with algae (with the exception for Colwellia), were consistently found on all three A. nosodum samples. Even more interesting, 9 of the 11 above mentioned genera containing polysaccharolytic isolates were predominant in this common core. The cultivable fraction of the bacterial community associated with A. nodosum is, thus, significantly enriched in macroalgal-polysaccharide-degrading bacteria and these bacteria seem important for the seaweed holobiont even though they are under-represented in alga-associated microbiome studies.
Keywords: Ascophyllum nodosum; Flavobacteriia; Gammaproteobacteria; agarase; algal polysaccharidase; alginate lyase; carrageenase; macroalgae.
Figures
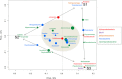
 ) and of the MAPD isolates belonging to these genera in the whole set of 78 MAPD isolates (
) and of the MAPD isolates belonging to these genera in the whole set of 78 MAPD isolates ( ); (B) Percentage proportions of MAPD isolates belonging to each MAPD-isolate-containing genus in the whole set of 78 MAPD isolates (
); (B) Percentage proportions of MAPD isolates belonging to each MAPD-isolate-containing genus in the whole set of 78 MAPD isolates ( ), with their activities on red (
), with their activities on red ( ) or brown seaweed galactans (
) or brown seaweed galactans ( ).
).
Similar articles
-
Discovering novel enzymes by functional screening of plurigenomic libraries from alga-associated Flavobacteriia and Gammaproteobacteria.Microbiol Res. 2016 May-Jun;186-187:52-61. doi: 10.1016/j.micres.2016.03.005. Epub 2016 Mar 24. Microbiol Res. 2016. PMID: 27242143
-
Evolutionary Evidence of Algal Polysaccharide Degradation Acquisition by Pseudoalteromonas carrageenovora 9T to Adapt to Macroalgal Niches.Front Microbiol. 2018 Nov 22;9:2740. doi: 10.3389/fmicb.2018.02740. eCollection 2018. Front Microbiol. 2018. PMID: 30524390 Free PMC article.
-
Microbial Population Changes in Decaying Ascophyllum nodosum Result in Macroalgal-Polysaccharide-Degrading Bacteria with Potential Applicability in Enzyme-Assisted Extraction Technologies.Mar Drugs. 2019 Mar 29;17(4):200. doi: 10.3390/md17040200. Mar Drugs. 2019. PMID: 30934874 Free PMC article.
-
Current knowledge and future perspectives of the use of seaweeds for livestock production and meat quality: a systematic review.J Anim Physiol Anim Nutr (Berl). 2021 Nov;105(6):1075-1102. doi: 10.1111/jpn.13509. Epub 2021 Mar 4. J Anim Physiol Anim Nutr (Berl). 2021. PMID: 33660883 Review.
-
Insights into Algal Polysaccharides: A Review of Their Structure, Depolymerases, and Metabolic Pathways.J Agric Food Chem. 2022 Feb 16;70(6):1749-1765. doi: 10.1021/acs.jafc.1c05365. Epub 2022 Feb 7. J Agric Food Chem. 2022. PMID: 35124966 Review.
Cited by
-
Enhanced algin oligosaccharide production through selective breeding and optimization of growth and degradation conditions in Cobetia sp. cqz5-12-M1.Sci Rep. 2024 Aug 22;14(1):19550. doi: 10.1038/s41598-024-70472-w. Sci Rep. 2024. PMID: 39174820 Free PMC article.
-
A red algal polysaccharide influences the multicellular development of the choanoflagellate Salpingoeca rosetta.bioRxiv [Preprint]. 2024 May 15:2024.05.14.594265. doi: 10.1101/2024.05.14.594265. bioRxiv. 2024. PMID: 38798503 Free PMC article. Preprint.
-
Maribacter halichondriae sp. nov., isolated from the marine sponge Halichondria panicea, displays features of a sponge-associated life style.Antonie Van Leeuwenhoek. 2024 Mar 15;117(1):56. doi: 10.1007/s10482-024-01950-4. Antonie Van Leeuwenhoek. 2024. PMID: 38489089 Free PMC article.
-
Metagenomic insights into the dynamic degradation of brown algal polysaccharides by kelp-associated microbiota.Appl Environ Microbiol. 2024 Feb 21;90(2):e0202523. doi: 10.1128/aem.02025-23. Epub 2024 Jan 23. Appl Environ Microbiol. 2024. PMID: 38259074 Free PMC article.
-
Three novel marine species of the genus Reichenbachiella exhibiting degradation of complex polysaccharides.Front Microbiol. 2023 Dec 14;14:1265676. doi: 10.3389/fmicb.2023.1265676. eCollection 2023. Front Microbiol. 2023. PMID: 38156005 Free PMC article.
References
-
- Akagawa-Matsushita M., Matsuo M., Koga Y., Yamasato K. (1992). Alteromonas atlantica sp. nov. and Alteromonas carrageenovora sp. nov., bacteria that decompose algal polysaccharides. Int. J. Syst. Bacteriol. 42, 621–627. 10.1099/00207713-42-4-621 - DOI
-
- Barbeyron T., Carpentier F., L'Haridon S., Schüler M., Michel G., Amann R. (2008). Description of maribacter forsetii sp. nov., a marine Flavobacteriaceae isolated from North Sea water, and emended description of the genus Maribacter. Int. J. Syst. Evol. Microbiol. 58, 790–797. 10.1099/ijs.0.65469-0 - DOI - PubMed
-
- Barbeyron T., L'Haridon S., Corre E., Kloareg B., Potin P. (2001). Zobellia galactanovorans gen. nov., sp. nov., a marine species of Flavobacteriaceae isolated from a red alga, and classification of [Cytophaga] uliginosa (ZoBell and Upham 1944) Reichenbach 1989 as Zobellia uliginosa gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 51, 985–997. 10.1099/00207713-51-3-985 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources


