Protein-protein docking: from interaction to interactome
- PMID: 25418159
- PMCID: PMC4213718
- DOI: 10.1016/j.bpj.2014.08.033
Protein-protein docking: from interaction to interactome
Abstract
The protein-protein docking problem is one of the focal points of activity in computational biophysics and structural biology. The three-dimensional structure of a protein-protein complex, generally, is more difficult to determine experimentally than the structure of an individual protein. Adequate computational techniques to model protein interactions are important because of the growing number of known protein structures, particularly in the context of structural genomics. Docking offers tools for fundamental studies of protein interactions and provides a structural basis for drug design. Protein-protein docking is the prediction of the structure of the complex, given the structures of the individual proteins. In the heart of the docking methodology is the notion of steric and physicochemical complementarity at the protein-protein interface. Originally, mostly high-resolution, experimentally determined (primarily by x-ray crystallography) protein structures were considered for docking. However, more recently, the focus has been shifting toward lower-resolution modeled structures. Docking approaches have to deal with the conformational changes between unbound and bound structures, as well as the inaccuracies of the interacting modeled structures, often in a high-throughput mode needed for modeling of large networks of protein interactions. The growing number of docking developers is engaged in the community-wide assessments of predictive methodologies. The development of more powerful and adequate docking approaches is facilitated by rapidly expanding information and data resources, growing computational capabilities, and a deeper understanding of the fundamental principles of protein interactions.
Figures
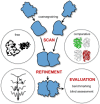


Similar articles
-
Predicting 3D structures of protein-protein complexes.Curr Pharm Biotechnol. 2008 Apr;9(2):57-66. doi: 10.2174/138920108783955209. Curr Pharm Biotechnol. 2008. PMID: 18393862 Review.
-
Modeling complexes of modeled proteins.Proteins. 2017 Mar;85(3):470-478. doi: 10.1002/prot.25183. Epub 2016 Oct 24. Proteins. 2017. PMID: 27701777 Free PMC article.
-
Across-proteome modeling of dimer structures for the bottom-up assembly of protein-protein interaction networks.BMC Bioinformatics. 2017 May 12;18(1):257. doi: 10.1186/s12859-017-1675-z. BMC Bioinformatics. 2017. PMID: 28499419 Free PMC article.
-
Simulated unbound structures for benchmarking of protein docking in the DOCKGROUND resource.BMC Bioinformatics. 2015 Jul 31;16(1):243. doi: 10.1186/s12859-015-0672-3. BMC Bioinformatics. 2015. PMID: 26227548 Free PMC article.
-
Low-resolution structural modeling of protein interactome.Curr Opin Struct Biol. 2013 Apr;23(2):198-205. doi: 10.1016/j.sbi.2012.12.003. Epub 2013 Jan 5. Curr Opin Struct Biol. 2013. PMID: 23294579 Free PMC article. Review.
Cited by
-
Future Perspective: Harnessing the Power of Artificial Intelligence in the Generation of New Peptide Drugs.Biomolecules. 2024 Oct 15;14(10):1303. doi: 10.3390/biom14101303. Biomolecules. 2024. PMID: 39456236 Free PMC article. Review.
-
Unified Sampling and Ranking for Protein Docking with DFMDock.bioRxiv [Preprint]. 2024 Sep 28:2024.09.27.615401. doi: 10.1101/2024.09.27.615401. bioRxiv. 2024. PMID: 39386449 Free PMC article. Preprint.
-
CAPRI-Q: The CAPRI resource evaluating the quality of predicted structures of protein complexes.J Mol Biol. 2024 Sep 1;436(17):168540. doi: 10.1016/j.jmb.2024.168540. Epub 2024 Mar 16. J Mol Biol. 2024. PMID: 39237205
-
Diffusion of proteins in crowded solutions studied by docking-based modeling.J Chem Phys. 2024 Sep 7;161(9):095101. doi: 10.1063/5.0220545. J Chem Phys. 2024. PMID: 39225532 Free PMC article.
-
Therapeutic Effects of Qingchang Tongluo Decoction on Intestinal Fibrosis in Crohn's Disease: Network Pharmacology, Molecular Docking and Experiment Validation.Drug Des Devel Ther. 2024 Jul 25;18:3269-3293. doi: 10.2147/DDDT.S458811. eCollection 2024. Drug Des Devel Ther. 2024. PMID: 39081706 Free PMC article.
References
-
- Platzer K.E.B., Momany F.A., Scheraga H.A. Conformational energy calculations of enzyme-substrate interactions. II. Computation of the binding energy for substrates in the active site of alpha-chymotrypsin. Int. J. Pept. Protein Res. 1972;4:201–219. - PubMed
-
- Wodak S.Y., Liu M.Y., Wyckoff H.W. The structure of cytidilyl(2′,5′)adenosine when bound to pancreatic ribonuclease S. J. Mol. Biol. 1977;116:855–875. - PubMed
-
- Pincus M.R., Scheraga H.A. Conformational energy calculations of enzyme-substrate and enzyme-inhibitor complexes of lysozyme. 2. Calculation of the structures of complexes with flexible enzyme. Macromolecules. 1979;12:633–644.
-
- Wodak S.J., Janin J. Computer analysis of protein-protein interaction. J. Mol. Biol. 1978;124:323–342. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources


