Genome-wide signatures of population bottlenecks and diversifying selection in European wolves
- PMID: 24346500
- PMCID: PMC3966127
- DOI: 10.1038/hdy.2013.122
Genome-wide signatures of population bottlenecks and diversifying selection in European wolves
Abstract
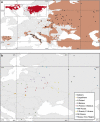
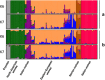
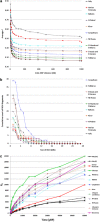
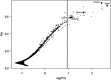
Genomic resources developed for domesticated species provide powerful tools for studying the evolutionary history of their wild relatives. Here we use 61K single-nucleotide polymorphisms (SNPs) evenly spaced throughout the canine nuclear genome to analyse evolutionary relationships among the three largest European populations of grey wolves in comparison with other populations worldwide, and investigate genome-wide effects of demographic bottlenecks and signatures of selection. European wolves have a discontinuous range, with large and connected populations in Eastern Europe and relatively smaller, isolated populations in Italy and the Iberian Peninsula. Our results suggest a continuous decline in wolf numbers in Europe since the Late Pleistocene, and long-term isolation and bottlenecks in the Italian and Iberian populations following their divergence from the Eastern European population. The Italian and Iberian populations have low genetic variability and high linkage disequilibrium, but relatively few autozygous segments across the genome. This last characteristic clearly distinguishes them from populations that underwent recent drastic demographic declines or founder events, and implies long-term bottlenecks in these two populations. Although genetic drift due to spatial isolation and bottlenecks seems to be a major evolutionary force diversifying the European populations, we detected 35 loci that are putatively under diversifying selection. Two of these loci flank the canine platelet-derived growth factor gene, which affects bone growth and may influence differences in body size between wolf populations. This study demonstrates the power of population genomics for identifying genetic signals of demographic bottlenecks and detecting signatures of directional selection in bottlenecked populations, despite their low background variability.
Figures


Similar articles
-
Genomic evidence for the Old divergence of Southern European wolf populations.Proc Biol Sci. 2020 Jul 29;287(1931):20201206. doi: 10.1098/rspb.2020.1206. Epub 2020 Jul 22. Proc Biol Sci. 2020. PMID: 32693716 Free PMC article.
-
Wolf population genetics in Europe: a systematic review, meta-analysis and suggestions for conservation and management.Biol Rev Camb Philos Soc. 2017 Aug;92(3):1601-1629. doi: 10.1111/brv.12298. Epub 2016 Sep 28. Biol Rev Camb Philos Soc. 2017. PMID: 27682639 Review.
-
Genetic variability of the grey wolf Canis lupus in the Caucasus in comparison with Europe and the Middle East: distinct or intermediary population?PLoS One. 2014 Apr 8;9(4):e93828. doi: 10.1371/journal.pone.0093828. eCollection 2014. PLoS One. 2014. PMID: 24714198 Free PMC article.
-
Howling from the past: historical phylogeography and diversity losses in European grey wolves.Proc Biol Sci. 2018 Aug 1;285(1884):20181148. doi: 10.1098/rspb.2018.1148. Proc Biol Sci. 2018. PMID: 30068681 Free PMC article.
-
Plant-based remedies for wolf bites and rituals against wolves in the Iberian Peninsula: Therapeutic opportunities and cultural values for the conservation of biocultural diversity.J Ethnopharmacol. 2017 Sep 14;209:124-139. doi: 10.1016/j.jep.2017.07.038. Epub 2017 Jul 27. J Ethnopharmacol. 2017. PMID: 28755969 Review.
Cited by
-
Imputation of ancient canid genomes reveals inbreeding history over the past 10,000 years.bioRxiv [Preprint]. 2024 Jul 3:2024.03.15.585179. doi: 10.1101/2024.03.15.585179. bioRxiv. 2024. PMID: 38903121 Free PMC article. Preprint.
-
Genetic Monitoring of Grey Wolves in Latvia Shows Adverse Reproductive and Social Consequences of Hunting.Biology (Basel). 2023 Sep 19;12(9):1255. doi: 10.3390/biology12091255. Biology (Basel). 2023. PMID: 37759654 Free PMC article.
-
Wolf genetic diversity compared across Europe using the yardstick method.Sci Rep. 2023 Aug 22;13(1):13727. doi: 10.1038/s41598-023-40834-x. Sci Rep. 2023. PMID: 37608038 Free PMC article.
-
Traces of Human-Mediated Selection in the Gene Pool of Red Deer Populations.Animals (Basel). 2023 Aug 4;13(15):2525. doi: 10.3390/ani13152525. Animals (Basel). 2023. PMID: 37570333 Free PMC article.
-
African wild dogs (Lycaon pictus) from the Kruger National Park, South Africa are currently not inbred but have low genomic diversity.Sci Rep. 2022 Sep 2;12(1):14979. doi: 10.1038/s41598-022-19025-7. Sci Rep. 2022. PMID: 36056068 Free PMC article.
References
-
- Allendorf FW, Hohenlohe PA, Luikart G. Genomics and the future of conservation genetics. Nat Rev Genet. 2010;11:697–709. - PubMed
-
- Altobello G.1921[Fauna of Abruzzo and Molise] Mammiferi 438–45.[In Italian].
-
- Alvarez RH, Kantarjian HM, Cortes JE. Biology of platelet-derived growth factor and its involvement in disease. Mayo Clin Proc. 2006;81:1241–1257. - PubMed
-
- Axelsson E, Ratnakuma A, Arendt ML, Maqbool K, Webster MT, Perloski M, et al. The genomic signature of dog domestication reveals adaptation to a starch-rich diet. Nature. 2013;495:360–364. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources


