Contrasting extracellular enzyme activities of particle-associated bacteria from distinct provinces of the North Atlantic Ocean
- PMID: 23248623
- PMCID: PMC3521168
- DOI: 10.3389/fmicb.2012.00425
Contrasting extracellular enzyme activities of particle-associated bacteria from distinct provinces of the North Atlantic Ocean
Abstract
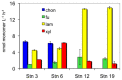
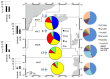
Microbial communities play a key role in the marine carbon cycle, processing much of phytoplankton-derived organic matter. The composition of these communities varies by depth, season, and location in the ocean; the functional consequences of these compositional variations for the carbon cycle are only beginning to be explored. We measured the abilities of microbial communities in the large-particle fraction (retained by a 10-μm pore-size cartridge filter) to enzymatically hydrolyze high molecular weight substrates, and therefore initiate carbon remineralization in four distinct oceanic provinces: the boreal polar (BPLR), the Arctic oceanic (ARCT), the North Atlantic drift (NADR), and the North Atlantic subtropical (NAST) provinces. Since we expected the large-particle fraction to include phytoplankton cells, we measured the hydrolysis of polysaccharide substrates (laminarin, fucoidan, xylan, and chondroitin sulfate) expected to be associated with phytoplankton. Hydrolysis rates and patterns clustered into two groups, the BPLR/ARCT and the NADR/NAST. All four substrates were hydrolyzed by the BPLR/ARCT communities; hydrolysis rates of individual substrate varied by factors of ca. 1-4. In contrast, chondroitin was not hydrolyzed in the NADR/NAST, and hydrolytic activity was dominated by laminarinase. Fluorescence in situ hybridization of the large-particle fraction post-incubation showed a substantial contribution (15-26%) of CF319a-positive cells (Bacteroidetes) to total DAPI-stainable cells. Concurrent studies of microbial community composition and of fosmids from these same stations also demonstrated similarities between BPLR and ARCT stations, which were distinct from the NADR/NAST stations. Together, these data support a picture of compositionally as well as functionally distinct communities across these oceanic provinces.
Keywords: biogeography; carbon cycling; extracellular enzymes; hydrolysis; particles-associated bacteria.
Figures
Similar articles
-
Genomic content of uncultured Bacteroidetes from contrasting oceanic provinces in the North Atlantic Ocean.Environ Microbiol. 2012 Jan;14(1):52-66. doi: 10.1111/j.1462-2920.2011.02555.x. Epub 2011 Sep 6. Environ Microbiol. 2012. PMID: 21895912
-
Distinct flavobacterial communities in contrasting water masses of the north Atlantic Ocean.ISME J. 2010 Apr;4(4):472-87. doi: 10.1038/ismej.2009.142. Epub 2010 Jan 7. ISME J. 2010. PMID: 20054356
-
Polysaccharide utilisation loci of Bacteroidetes from two contrasting open ocean sites in the North Atlantic.Environ Microbiol. 2016 Dec;18(12):4456-4470. doi: 10.1111/1462-2920.13429. Epub 2016 Jul 18. Environ Microbiol. 2016. PMID: 27348854
-
Extensive Microbial Processing of Polysaccharides in the South Pacific Gyre via Selfish Uptake and Extracellular Hydrolysis.Front Microbiol. 2020 Dec 18;11:583158. doi: 10.3389/fmicb.2020.583158. eCollection 2020. Front Microbiol. 2020. PMID: 33391202 Free PMC article.
-
Microbial extracellular enzymes and the marine carbon cycle.Ann Rev Mar Sci. 2011;3:401-25. doi: 10.1146/annurev-marine-120709-142731. Ann Rev Mar Sci. 2011. PMID: 21329211 Review.
Cited by
-
Strong chemotaxis by marine bacteria towards polysaccharides is enhanced by the abundant organosulfur compound DMSP.Nat Commun. 2023 Dec 6;14(1):8080. doi: 10.1038/s41467-023-43143-z. Nat Commun. 2023. PMID: 38057294 Free PMC article.
-
Seasonal Dynamics in Carbon Cycling of Marine Bacterioplankton Are Lifestyle Dependent.Front Microbiol. 2022 Jul 5;13:834675. doi: 10.3389/fmicb.2022.834675. eCollection 2022. Front Microbiol. 2022. PMID: 36212867 Free PMC article.
-
Marine Group-II archaea dominate particle-attached as well as free-living archaeal assemblages in the surface waters of Kongsfjorden, Svalbard, Arctic Ocean.Antonie Van Leeuwenhoek. 2021 May;114(5):633-647. doi: 10.1007/s10482-021-01547-1. Epub 2021 Mar 10. Antonie Van Leeuwenhoek. 2021. PMID: 33694023 Free PMC article.
-
How do Planktonic Particle Collection Methods Affect Bacterial Diversity Estimates and Community Composition in Oligo-, Meso- and Eutrophic Lakes?Front Microbiol. 2020 Dec 4;11:593589. doi: 10.3389/fmicb.2020.593589. eCollection 2020. Front Microbiol. 2020. PMID: 33343534 Free PMC article.
-
Depth-Dependent Variables Shape Community Structure and Functionality in the Prince Edward Islands.Microb Ecol. 2021 Feb;81(2):396-409. doi: 10.1007/s00248-020-01589-4. Epub 2020 Sep 15. Microb Ecol. 2021. PMID: 32935183
References
-
- Alderkamp A.-C., Van Rijssel M., Bolhuis H. (2007). Characterization of marine bacteria and the activity of their enzyme systems involved in degradation of the algal storage glucan laminarin. FEMS Microbiol. Ecol. 59 108–117 - PubMed
-
- Alonso-Saez L., Sanchez O., Gasol J. M., Balague V., Pedros-Alio C. (2008). Winter-to-summer changes in the composition and single-cell activity of near-surface Arctic prokaryotes. Environ. Microbiol. 10 2444–2454 - PubMed
-
- Amann R., Fuchs B. (2008). Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nat. Rev. Microbiol. 6 339–348 - PubMed
LinkOut - more resources
Full Text Sources


