Epigenetic mechanisms of mouse interstrain variability in genotoxicity of the environmental toxicant 1,3-butadiene
- PMID: 21602187
- PMCID: PMC3155089
- DOI: 10.1093/toxsci/kfr133
Epigenetic mechanisms of mouse interstrain variability in genotoxicity of the environmental toxicant 1,3-butadiene
Abstract
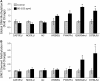
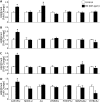
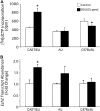
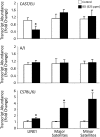
1,3-Butadiene (BD) is a common environmental contaminant classified as "carcinogenic to humans." Formation of BD-induced DNA adducts plays a major role in its carcinogenicity. BD is also an epigenotoxic agent (i.e., it affects DNA and histone methylation in the liver). We used a panel of genetically diverse inbred mice (NOD/LtJ, CAST/EiJ, A/J, WSB/EiJ, PWK/PhJ, C57BL/6J, and 129S1/SvImJ) to assess whether BD-induced genotoxic and epigenotoxic events may be subject to interstrain differences. Mice (male, 7 weeks) were exposed via inhalation to 0 or 625 ppm BD for 6 h/day and 5 days/week for 2 weeks and liver BD-DNA adducts, epigenetic alterations, and liver toxicity were assessed. N-7-(2,3,4-trihydroxybut-1-yl)-guanine adducts were detected in all strains after exposure, yet BD-induced DNA damage in CAST/EiJ mice was two to three times lower. Epigenetic effects of BD were most prominent in C57BL/6J mice where loss of global DNA methylation and loss of trimethylation of histone H3 lysine 9, histone H3 lysine 27, and histone H4 lysine 20, accompanied by dysregulation of liver gene expression indicative of hepatotoxicity, were found. Interestingly, we observed an increase in histone methylation in the absence of changes in gene expression and DNA methylation in CAST/EiJ strain. We hypothesized that mitigated genotoxicity of BD in CAST/EiJ mice may be due to chromatin condensation. Indeed, we show that in response to BD exposure, chromatin condensation occurs in CAST/EiJ, whereas the opposite effect is observed in C57BL/6J mice. These findings demonstrate that interstrain susceptibility to genotoxicity by a well-known environmental carcinogen may be due to strain-specific epigenetic events in response to the exposure.
Figures
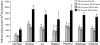

Similar articles
-
Epigenetic events determine tissue-specific toxicity of inhalational exposure to the genotoxic chemical 1,3-butadiene in male C57BL/6J mice.Toxicol Sci. 2014 Dec;142(2):375-84. doi: 10.1093/toxsci/kfu191. Epub 2014 Sep 18. Toxicol Sci. 2014. PMID: 25237060 Free PMC article.
-
Epigenetic alterations in liver of C57BL/6J mice after short-term inhalational exposure to 1,3-butadiene.Environ Health Perspect. 2011 May;119(5):635-40. doi: 10.1289/ehp.1002910. Epub 2010 Dec 13. Environ Health Perspect. 2011. PMID: 21147608 Free PMC article.
-
Sex-specific differences in genotoxic and epigenetic effects of 1,3-butadiene among mouse tissues.Arch Toxicol. 2019 Mar;93(3):791-800. doi: 10.1007/s00204-018-2374-x. Epub 2018 Dec 14. Arch Toxicol. 2019. PMID: 30552462 Free PMC article.
-
A review of the genetic and related effects of 1,3-butadiene in rodents and humans.Mutat Res. 2000 Oct;463(3):181-213. doi: 10.1016/s1383-5742(00)00056-9. Mutat Res. 2000. PMID: 11018742 Review.
-
Epidemiological and mechanistic data suggest that 1,3-butadiene will not be carcinogenic to humans at exposures likely to be encountered in the environment or workplace.Carcinogenesis. 1995 Feb;16(2):165-71. doi: 10.1093/carcin/16.2.165. Carcinogenesis. 1995. PMID: 7859344 Review.
Cited by
-
Stem cell transcriptional profiles from mouse subspecies reveal cis-regulatory evolution at translation genes.Heredity (Edinb). 2024 Nov;133(5):308-316. doi: 10.1038/s41437-024-00715-z. Epub 2024 Aug 20. Heredity (Edinb). 2024. PMID: 39164520 Free PMC article.
-
Inter-strain variability in responses to a single administration of the cannabidiol-rich cannabis extract in mice.Food Chem Toxicol. 2024 Oct;192:114909. doi: 10.1016/j.fct.2024.114909. Epub 2024 Aug 9. Food Chem Toxicol. 2024. PMID: 39128689
-
Model systems and organisms for addressing inter- and intra-species variability in risk assessment.Regul Toxicol Pharmacol. 2022 Jul;132:105197. doi: 10.1016/j.yrtph.2022.105197. Epub 2022 May 28. Regul Toxicol Pharmacol. 2022. PMID: 35636685 Free PMC article. Review.
-
Characterization of population variability of 1,3-butadiene derived protein adducts in humans and mice.Regul Toxicol Pharmacol. 2022 Jul;132:105171. doi: 10.1016/j.yrtph.2022.105171. Epub 2022 Apr 22. Regul Toxicol Pharmacol. 2022. PMID: 35469930 Free PMC article.
-
Folate metabolism modifies chromosomal damage induced by 1,3-butadiene: results from a match-up study in China and in vitro experiments.Genes Environ. 2021 Oct 9;43(1):44. doi: 10.1186/s41021-021-00217-y. Genes Environ. 2021. PMID: 34627392 Free PMC article.
References
-
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in polycomb-group silencing. Science. 2002;298:1039–1043. - PubMed
-
- Chen RZ, Pettersson U, Beard C, Jackson-Grusby L, Jaenisch R. DNA hypomethylation leads to elevated mutation rates. Nature. 1998;395:89–93. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous


