Erythrophore cell response to food-associated pathogenic bacteria: implications for detection
- PMID: 21261862
- PMCID: PMC3815249
- DOI: 10.1111/j.1751-7915.2008.00045.x
Erythrophore cell response to food-associated pathogenic bacteria: implications for detection
Abstract
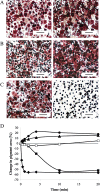
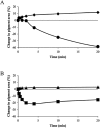
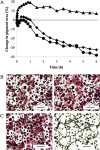
Cell-based biosensors have been proposed for use as function-based detectors of toxic agents. We report the use of Betta splendens chromatophore cells, specifically erythrophore cells, for detection of food-associated pathogenic bacteria. Evaluation of erythrophore cell response, using Bacillus spp., has revealed that this response can distinguish pathogenic Bacillus cereus from a non-pathogenic B. cereus ΔplcR deletion mutant and a non-pathogenic Bacillus subtilis. Erythrophore cells were exposed to Salmonella enteritidis, Clostridium perfringens and Clostridium botulinum. Each bacterial pathogen elicited a response from erythrophore cells that was distinguished from the corresponding bacterial growth medium, and this observed response was unique for each bacterial pathogen. These findings suggest that erythrophore cell response has potential for use as a biosensor in the detection and toxicity assessment for food-associated pathogenic bacteria.
© 2008 The Authors. Journal compilation © 2008 Society for Applied Microbiology and Blackwell Publishing Ltd.
Figures
Similar articles
-
Trends and opportunities in food pathogen detection.Anal Bioanal Chem. 2008 May;391(2):451-4. doi: 10.1007/s00216-008-1886-2. Epub 2008 Mar 18. Anal Bioanal Chem. 2008. PMID: 18347781 Free PMC article. No abstract available.
-
Electroanalytical biosensors and their potential for food pathogen and toxin detection.Anal Bioanal Chem. 2008 May;391(2):455-71. doi: 10.1007/s00216-008-1876-4. Epub 2008 Feb 17. Anal Bioanal Chem. 2008. PMID: 18283441 Review.
-
Electrical/electrochemical impedance for rapid detection of foodborne pathogenic bacteria.Biotechnol Adv. 2008 Mar-Apr;26(2):135-50. doi: 10.1016/j.biotechadv.2007.10.003. Epub 2007 Nov 12. Biotechnol Adv. 2008. PMID: 18155870 Review.
-
Nanomaterial-based biosensors for sensing key foodborne pathogens: Advances from recent decades.Compr Rev Food Sci Food Saf. 2020 Jul;19(4):1465-1487. doi: 10.1111/1541-4337.12576. Epub 2020 Jun 7. Compr Rev Food Sci Food Saf. 2020. PMID: 33337098 Review.
-
Cell-based biosensor for rapid screening of pathogens and toxins.Biosens Bioelectron. 2010 Sep 15;26(1):99-106. doi: 10.1016/j.bios.2010.05.020. Epub 2010 May 19. Biosens Bioelectron. 2010. PMID: 20570502
Cited by
-
Environmental Microbiology meets Microbial Biotechnology.Microb Biotechnol. 2008 Nov;1(6):443-5. doi: 10.1111/j.1751-7915.2008.00068.x. Microb Biotechnol. 2008. PMID: 21261865 Free PMC article. No abstract available.
-
Potential of the melanophore pigment response for detection of bacterial toxicity.Appl Environ Microbiol. 2010 Dec;76(24):8243-6. doi: 10.1128/AEM.01241-10. Epub 2010 Oct 15. Appl Environ Microbiol. 2010. PMID: 20952639 Free PMC article.
References
-
- Agaisse H., Gominet M., Økstad O.A., Kolsto A.B., Lereclus D. PlcR is a pleiotropic regulator of extracellular virulence factor gene expression in Bacillus thuringiensis. Mol Microbiol. 1999;32:1043–1053. - PubMed
-
- Batt C.A. Materials science: food pathogen detection. Science. 2007;104:6404–6405. - PubMed
-
- Buzby J.C., Roberts T. Economic costs and trade impacts of microbial foodborne illness. World Health Stat Q. 1997;50:57–66. - PubMed
-
- Cady N.C., Stelick S., Kunnavakkam M.V., Batt C.A. Real‐time PCR detection of Listeria monocytogenes using an integrated microFLUIDICS platform. Sens Actuators B. 2005;107:332–341.
-
- Danosky T.R., McFadden P.N. Biosensors based on the activities of living, naturally pigmented cells: digital image processing of the dynamics of fish melanophores. Biosens Bioelectron. 1997;12:925–936.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous


