Base-excision repair of oxidative DNA damage
- PMID: 17581577
- PMCID: PMC2896554
- DOI: 10.1038/nature05978
Base-excision repair of oxidative DNA damage
Abstract
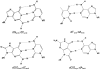
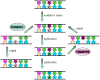
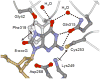
Maintaining the chemical integrity of DNA in the face of assault by oxidizing agents is a constant challenge for living organisms. Base-excision repair has an important role in preventing mutations associated with a common product of oxidative damage to DNA, 8-oxoguanine. Recent structural studies have shown that 8-oxoguanine DNA glycosylases use an intricate series of steps to locate and excise 8-oxoguanine lesions efficiently against a high background of undamaged bases. The importance of preventing mutations associated with 8-oxoguanine is shown by a direct association between defects in the DNA glycosylase MUTYH and colorectal cancer. The properties of other guanine oxidation products and the associated DNA glycosylases that remove them are now also being revealed.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

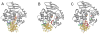
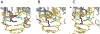
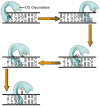
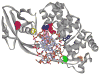

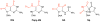
Similar articles
-
The role of the N-terminal domain of human apurinic/apyrimidinic endonuclease 1, APE1, in DNA glycosylase stimulation.DNA Repair (Amst). 2018 Apr;64:10-25. doi: 10.1016/j.dnarep.2018.02.001. Epub 2018 Feb 11. DNA Repair (Amst). 2018. PMID: 29475157
-
The DNA repair enzyme MUTYH potentiates cytotoxicity of the alkylating agent MNNG by interacting with abasic sites.J Biol Chem. 2020 Mar 13;295(11):3692-3707. doi: 10.1074/jbc.RA119.010497. Epub 2020 Jan 30. J Biol Chem. 2020. PMID: 32001618 Free PMC article.
-
Excision of 8-oxoguanine from methylated CpG dinucleotides by human 8-oxoguanine DNA glycosylase.FEBS Lett. 2013 Sep 17;587(18):3129-34. doi: 10.1016/j.febslet.2013.08.008. Epub 2013 Aug 13. FEBS Lett. 2013. PMID: 23954288
-
UV-DDB as a General Sensor of DNA Damage in Chromatin: Multifaceted Approaches to Assess Its Direct Role in Base Excision Repair.Int J Mol Sci. 2023 Jun 15;24(12):10168. doi: 10.3390/ijms241210168. Int J Mol Sci. 2023. PMID: 37373320 Free PMC article. Review.
-
Repair of 8-oxoG:A mismatches by the MUTYH glycosylase: Mechanism, metals and medicine.Free Radic Biol Med. 2017 Jun;107:202-215. doi: 10.1016/j.freeradbiomed.2017.01.008. Epub 2017 Jan 10. Free Radic Biol Med. 2017. PMID: 28087410 Free PMC article. Review.
Cited by
-
Human 8-oxoguanine glycosylase OGG1 binds nucleosome at the dsDNA ends and the super-helical locations.Commun Biol. 2024 Sep 28;7(1):1202. doi: 10.1038/s42003-024-06919-7. Commun Biol. 2024. PMID: 39341999 Free PMC article.
-
DNA damage response in breast cancer and its significant role in guiding novel precise therapies.Biomark Res. 2024 Sep 27;12(1):111. doi: 10.1186/s40364-024-00653-2. Biomark Res. 2024. PMID: 39334297 Free PMC article. Review.
-
Innate immune cells in tumor microenvironment: A new frontier in cancer immunotherapy.iScience. 2024 Aug 17;27(9):110750. doi: 10.1016/j.isci.2024.110750. eCollection 2024 Sep 20. iScience. 2024. PMID: 39280627 Free PMC article. Review.
-
8-OxodG: A Potential Biomarker for Chronic Oxidative Stress Induced by High-LET Radiation.DNA (Basel). 2024 Sep;4(3):221-238. doi: 10.3390/dna4030015. Epub 2024 Aug 1. DNA (Basel). 2024. PMID: 39268222 Free PMC article.
-
CO2 protects cells from iron-Fenton oxidative DNA damage in E. coli and humans.bioRxiv [Preprint]. 2024 Aug 26:2024.08.26.609766. doi: 10.1101/2024.08.26.609766. bioRxiv. 2024. Update in: Proc Natl Acad Sci U S A. 2024 Dec 3;121(49):e2419175121. doi: 10.1073/pnas.2419175121 PMID: 39253463 Free PMC article. Updated. Preprint.
References
-
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. - PubMed
-
- Friedberg EC. DNA Damage and Repair. Nature. 2003;421:436–440. - PubMed
-
- Pfeifer GP, et al. Tobacoo smoke carcinogens, DNA Damage and p53 mutations in smoking-associated cancers. Oncogene. 2002;21:7435–51. - PubMed
-
- Friedberg EC. Inroads into base excision repair II. The discovery of the DNA glycosylases. DNA Repair. 2004;3:1531–1536. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials


