Phylogeography and genetic ancestry of tigers (Panthera tigris)
- PMID: 15583716
- PMCID: PMC534810
- DOI: 10.1371/journal.pbio.0020442
Phylogeography and genetic ancestry of tigers (Panthera tigris)
Abstract
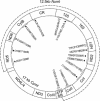
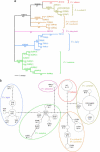
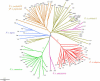
Eight traditional subspecies of tiger (Panthera tigris),of which three recently became extinct, are commonly recognized on the basis of geographic isolation and morphological characteristics. To investigate the species' evolutionary history and to establish objective methods for subspecies recognition, voucher specimens of blood, skin, hair, and/or skin biopsies from 134 tigers with verified geographic origins or heritage across the whole distribution range were examined for three molecular markers: (1) 4.0 kb of mitochondrial DNA (mtDNA) sequence; (2) allele variation in the nuclear major histocompatibility complex class II DRB gene; and (3) composite nuclear microsatellite genotypes based on 30 loci. Relatively low genetic variation with mtDNA,DRB,and microsatellite loci was found, but significant population subdivision was nonetheless apparent among five living subspecies. In addition, a distinct partition of the Indochinese subspecies P. t. corbetti in to northern Indochinese and Malayan Peninsula populations was discovered. Population genetic structure would suggest recognition of six taxonomic units or subspecies: (1) Amur tiger P. t. altaica; (2) northern Indochinese tiger P. t. corbetti; (3) South China tiger P. t. amoyensis; (4) Malayan tiger P. t. jacksoni, named for the tiger conservationist Peter Jackson; (5) Sumatran tiger P. t. sumatrae; and (6) Bengal tiger P. t. tigris. The proposed South China tiger lineage is tentative due to limited sampling. The age of the most recent common ancestor for tiger mtDNA was estimated to be 72,000-108,000 y, relatively younger than some other Panthera species. A combination of population expansions, reduced gene flow, and genetic drift following the last genetic diminution, and the recent anthropogenic range contraction, have led to the distinct genetic partitions. These results provide an explicit basis for subspecies recognition and will lead to the improved management and conservation of these recently isolated but distinct geographic populations of tigers.
Conflict of interest statement
The authors have declared that no conflicts of interests exist.
Figures

Similar articles
-
Subspecies genetic assignments of worldwide captive tigers increase conservation value of captive populations.Curr Biol. 2008 Apr 22;18(8):592-6. doi: 10.1016/j.cub.2008.03.053. Curr Biol. 2008. PMID: 18424146
-
Sorting Out the Genetic Background of the Last Surviving South China Tigers.J Hered. 2019 Oct 10;110(6):641-650. doi: 10.1093/jhered/esz034. J Hered. 2019. PMID: 31102441
-
Genetic ancestry of the extinct Javan and Bali tigers.J Hered. 2015 May-Jun;106(3):247-57. doi: 10.1093/jhered/esv002. Epub 2015 Mar 8. J Hered. 2015. PMID: 25754539 Free PMC article.
-
Securing a future for wild Indochinese tigers: Transforming tiger vacuums into tiger source sites.Integr Zool. 2010 Dec;5(4):324-334. doi: 10.1111/j.1749-4877.2010.00220.x. Integr Zool. 2010. PMID: 21392350 Review.
-
Sumatran tiger (Panthera tigris sumatrae): a review of conservation status.Integr Zool. 2010 Dec;5(4):313-323. doi: 10.1111/j.1749-4877.2010.00219.x. Integr Zool. 2010. PMID: 21392349 Review.
Cited by
-
Insights for the Captive Management of South China Tigers Based on a Large-Scale Genetic Survey.Genes (Basel). 2024 Mar 24;15(4):398. doi: 10.3390/genes15040398. Genes (Basel). 2024. PMID: 38674333 Free PMC article.
-
The genetic status and rescue measure for a geographically isolated population of Amur tigers.Sci Rep. 2024 Apr 6;14(1):8088. doi: 10.1038/s41598-024-58746-9. Sci Rep. 2024. PMID: 38582794 Free PMC article.
-
A Genetic Tool to Identify Predators Responsible for Livestock Attacks in South America and Recommendations for Human-Wildlife Conflict Mitigation.Animals (Basel). 2024 Mar 8;14(6):838. doi: 10.3390/ani14060838. Animals (Basel). 2024. PMID: 38539936 Free PMC article.
-
Acknowledging more biodiversity without more species.Proc Natl Acad Sci U S A. 2023 Oct 3;120(40):e2302424120. doi: 10.1073/pnas.2302424120. Epub 2023 Sep 25. Proc Natl Acad Sci U S A. 2023. PMID: 37748058 Free PMC article.
-
High-throughput DNA metabarcoding for determining the gut microbiome of captive critically endangered Malayan tiger (Pantheratigrisjacksoni) during fasting.Biodivers Data J. 2023 Sep 5;11:e104757. doi: 10.3897/BDJ.11.e104757. eCollection 2023. Biodivers Data J. 2023. PMID: 37711366 Free PMC article.
References
-
- Ambrose SH. Late Pleistocene human population bottlenecks, volcanic winter, and differentiation of modern humans. J Hum Evol. 1998;34:623–651. - PubMed
-
- Avise JC, Ball RM. Principles of genealogical concordance in species concepts and biological taxonomy. Oxford Surv Evol Biol. 1990;7:45–67.
-
- Bodmer JG, Marsh SGE, Albert ED, Bodmer WF, Bontrop RE, et al. Nomenclature for factors of the HLA system, 1998. Tissue Antigens. 1999;53:407–446. - PubMed
-
- Clement M, Posada D, Crandall KA. TCS: A computer program to estimate gene genealogies. Mol Ecol. 2000;9:1657–1659. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources


