Abstract
Collectively, genes encoding subunits of the SWI/SNF (BAF) chromatin remodeling complex are mutated in 20% of all human cancers, with the SMARCA4 (BRG1) subunit being one of the most frequently mutated. The SWI/SNF complex modulates chromatin remodeling through the activity of two mutually exclusive catalytic subunits, SMARCA4 and SMARCA2 (BRM). Here, we show that a SMARCA2-containing residual SWI/SNF complex underlies the oncogenic activity of SMARCA4 mutant cancers. We demonstrate that a residual SWI/SNF complex exists in SMARCA4 mutant cell lines and plays essential roles in cellular proliferation. Further, using data from loss-of-function screening of 165 cancer cell lines, we identify SMARCA2 as an essential gene in SMARCA4 mutant cancer cell lines. Mechanistically, we reveal that Smarca4 inactivation leads to greater incorporation of the nonessential SMARCA2 subunit into the SWI/SNF complex. Collectively, these results reveal a role for SMARCA2 in oncogenesis caused by SMARCA4 loss and identify the ATPase and bromodomain-containing SMARCA2 as a potential therapeutic target in these cancers.
INTRODUCTION
Growing evidence indicates that subunits of the SWI/SNF (BAF) complex serve essential roles in the initiation and progression of cancer. At least eight genes that encode SWI/SNF subunits are frequently mutated in a variety of different cancers (1–5). The SMARCA4 (BRG1) subunit is mutated in 10 to 35% of non-small-cell lung carcinoma, 15% of Burkitt's lymphoma, 5 to 10% of childhood medulloblastoma, and occasionally in pancreatic adenocarcinoma, ovarian clear cell carcinoma, and melanoma (2, 6–10). SMARCA4 has been validated as a bona fide tumor suppressor, as haploinsufficient mice are tumor prone (11–13). Other SWI/SNF subunits also have potent tumor suppressor functions. Recently, a comprehensive analysis of whole-exome and whole-genome sequencing studies revealed that collectively subunits of the SWI/SNF complex are specifically mutated in one-fifth of all human cancers (2). Given that SMARCA4 is one of the most broadly mutated subunits (3), developing an understanding of the mechanisms by which mutation of SMARCA4 drives cancer and of the vulnerabilities created carries major disease relevance.
A catalytic ATPase subunit in the SWI/SNF complex, SMARCA4, has been shown to mediate nucleosome repositioning and to regulate transcription of its targets. SMARCA2 (BRM), a homolog 75% identical to SMARCA4, is similarly capable of regulating chromatin structure and is mutually exclusive of SMARCA4 in the SWI/SNF complex (14–17). These subunits function together with other core subunits, which include SMARCC1, SMARCC2, and SMARCB1, as well as with a number of variant lineage-restricted subunits, and collectively contribute to the control of cell fate and lineage specification (18–20). However, SMARCA4 and SMARCA2 have important differences in expression and function. In human and mouse tissues, SMARCA4 and SMARCA2 are often expressed at different stages of development and in distinct cell and tissue types (21, 22). Homozygous inactivation of Smarca4 in mice leads to early embryonic lethality, whereas SMARCA2-deficient mice are viable and survive into adulthood (11, 23).
While SMARCA4 has emerged as a critical tumor suppressor, the mechanisms by which its mutation contributes to tumorigenesis, and whether its mutation creates cellular vulnerabilities, have been unknown. Given that at least eight SWI/SNF subunits are recurrently mutated in cancer, one possibility is that each of these mutations inactivates the SWI/SNF complex, resulting in mutational equivalency. However, based upon distinct associated cancer spectra and distinct mutational phenotypes in mice, we hypothesize that cancer driven by SWI/SNF mutations results from aberrant activity of the remaining subunits, which assemble into a residual complex. Indeed, we have previously shown that a residual SWI/SNF complex is essential in SMARCB1 mutant rhabdoid tumors (24). Here, we evaluate the role of residual SWI/SNF complexes in SMARCA4 mutant cancers.
MATERIALS AND METHODS
Cell culture.
NCI-H1299 (CRL-5803) and A549 (CCL-185) cell lines were purchased from the ATCC. NCI-H2122, H460, and HCC-827 were obtained from Jeffrey Shapiro's laboratory at Dana-Farber Cancer Institute. Derivation and manipulation of mouse embryonic fibroblasts (MEFs) were described previously (25). Transduced cells were selected in puromycin for 48 to 72 h before counting and seeding for colony formation assays. To evaluate colony-forming ability, cells were seeded at low density and incubated under standard conditions for 10 to 14 days before staining with crystal violet staining solution (0.05% crystal violet, 1% formaldehyde, 1% phosphate-buffered saline [PBS], 1% methanol) for 20 min.
shRNA-mediated knockdown of SWI/SNF subunits.
Doxycycline-inducible short hairpin RNAs (shRNAs) targeting SMARCB1 (V3LHS_367694, ACCAGTGTGACCCTGTTAA; V3LHS_367696, AGACAGCAGATCGAGTCCT; Open Biosystems), cloned into the pTRIPZ vector (RHS4750; Open Biosystems) or nonsilencing control shRNA (pTRIPZ-NS; RHS4743; Open Biosystems), were transduced into SMARCA4 mutant cell lines (NCI-H1299 and A549) using lentivirus. Expression was initiated by the addition of doxycycline (2.5 μg/ml; 631311; Clontech). Cell medium was replaced with fresh medium containing doxycycline every other day. shRNAs targeting SMARCA2 (TRCN0000358828, GGCCATCGAAGACGGCAATTT; TRCN0000330445, CTATATCATCATCGTCTATAA) and a nonsilencing control (TRCN0000072240, TCGTATTACAACGTCGTGACT) were obtained from the RNA interference (RNAi) screening facility at the Dana-Farber Cancer Institute and transduced into SMARCA4 mutant cell lines (NCI-H1299 and A549) and SMARCA4 wild-type cell lines (NCI-2122, NCI-H460, and HCC827). SMARCA2 and nonsilencing control shRNAs are in the pLKO lentiviral expression vector backbone.
Project Achilles.
Data from Project Achilles were used to identify differentially essential genes in SMARCA4 mutant cell lines. Project Achilles is a systematic approach aimed at determining cancer-specific vulnerabilities in cancer cell lines using genome-scale shRNA screens (http://www.broadinstitute.org/achilles) (26). We used the Achilles version 2.4 data set, consisting of 216 cell lines that were screened according to an unpublished method. Briefly, for the Achilles screens, cells were transduced with pooled shRNA libraries containing 54,020 shRNAs targeting 11,194 genes and propagated for at least 16 doublings. The abundance of remaining individual shRNAs was determined at the endpoint relative to an initial reference pool using next-generation sequencing. The subsequent analysis of Achilles data included the following steps. (i) Gene-level shRNA scores were derived from individual shRNA scores using ATARiS. ATARiS is a computational method that enriches for related phenotypic effects caused by individual shRNAs (27). (ii) The gene mutation status for SMARCA4 in cancer cell lines was next determined using information from the Cancer Cell Line Encyclopedia (www.broadinstitute.org/ccle) and prior publications (Table 1). Cell lines without hybrid capture sequencing data were removed from the analysis; thus, 165 of the 216 cell lines were used in SMARCA4 mutant and SMARCA4 wild-type comparisons. These data were used to generate a classifier file that was used in subsequent comparisons. (iii) Differentially essential genes were identified using the GenePattern module PARIS, which ranked ATARiS values based on the mutation status of the cell lines using the mutual information-based metric RNMI (rescaled normalized mutual information) (unpublished data; the PARIS GenePattern module will be available at http://www.broadinstitute.org/cancer/software/genepattern).
TABLE 1.
Cell lines with inactivating SMARCA4 mutations used in vulnerability screen
| Cell line | Tissue | SMARCA4 protein change | Variant classification | Heterozygosity |
|---|---|---|---|---|
| A549a | Lung | p.L728fs | Frame shift deletion | Homozygous |
| HEC1A | Endometrium | p.V268fs | Frame shift insertion | Heterozygous |
| NCIH23a | Lung | p.1598_1599KE>Na | Nonsense mutation | Homozygous |
| NCIH661a | Lung | p.G1159fs | Frame shift deletion | Homozygous |
| NCIH1299a | Lung | p.Tyr560fs | Frame shift | Homozygous |
| JHOC5b | Ovary | Large focal deletion | Deletion | Homozygous |
| SK-MEL-5b | Skin | p.F1053fs | Frame shift | Heterozygous |
| TYKNU | Ovary | Large focal deletion | Deletion | Homozygous |
Immunoblots and coimmunoprecipitation experiments.
Whole-cell extracts for immunoblotting were prepared by incubating cells on ice in 1% NP-40 lysis buffer (50 mM Tris-HCl, pH 7.4, 5 mM EDTA, 12% glycerol, 50 mM NaCl, 1% NP-40) plus protease inhibitors (11836170001; Complete, Mini, EDTA-free; Roche) for 30 min. Supernatants were collected following centrifugation (10 min at 17,900 × g) at 4°C in a Legend 17R microcentrifuge (ClickSeal rotor 75003424). Protein concentrations were determined using the Bradford reagent (Bio-Rad). SDS-polyacrylamide gel electrophoresis was used to separate proteins, which were subsequently transferred to polyvinylidene difluoride (PVDF) membranes (Millipore). Antibodies used for immunoblotting included SMARCC1 (9746; Santa Cruz), ARID1A (A301-041A; Bethyl Laboratories), PBRM1 (A301-591A; Bethyl Laboratories), SMARCA4 (17796; Santa Cruz), SMARCC2 (A301-039A; Bethyl Laboratories), SMARCD1 (A301-595A; Bethyl Laboratories), SMARCE1 (A300-810A; Bethyl Laboratories), SMARCA2 (6889; Cell Signaling Technology), ACTL6A (A301-391A; Bethyl Laboratories), and actin (5125; Cell Signaling Technology) antibodies. Adobe Photoshop was used to prepare digital images (cropping and brightness/contrast) for publication.
Nuclear extracts for immunoprecipitation were prepared using the NE-PER nuclear and cytoplasmic extraction kit (78833; Thermo Scientific). Nuclear extracts (125 μg) were incubated in low-salt IP buffer (50 mM Tris-HCl [pH 7.5], 1 mM EDTA, 10% glycerol, 100 mM NaCl, 0.05% Tween 20, and protease inhibitors; the total volume was 300 μl) with antibodies (SMARCC1, 9746; Santa Cruz; SMARCA4, 17796; Santa Cruz; IgG, 2028; Santa Cruz) overnight at 4°C. Protein G Dynabeads (40 μl; 10004D; Life Technologies) were added and incubated at 4°C for 3.5 h. Beads were then washed twice with low-salt IP buffer and once with high-salt IP buffer (50 mM Tris-HCl [pH 7.5], 1 mM EDTA, 10% glycerol, 250 mM NaCl, 0.05% Tween 20). The beads were then resuspended in reducing SDS gel loading buffer.
Gene expression data.
Whole-genome expression data from SMARCA4-deficient and wild-type mouse embryonic fibroblasts was generated as previously described (28). Robust multiarray (RMA)-normalized expression values for Smarca2 in these samples were evaluated using Prism data analysis software. The P value was calculated using an unpaired t test with Welch's correction.
RESULTS
Residual SWI/SNF complexes are present in SMARCA4 mutant cell lines.
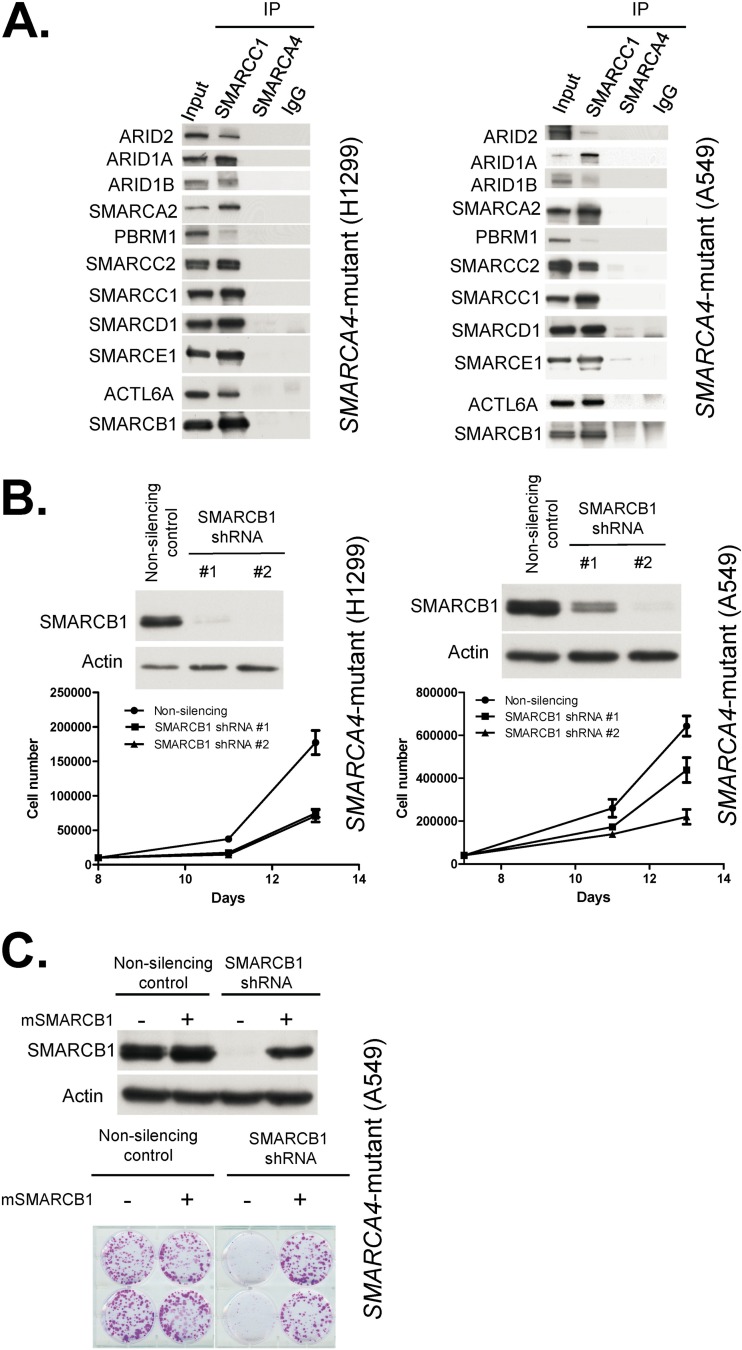
As SMARCA4 is widely mutated in cancer and is an essential and core ATPase of the SWI/SNF complex, we sought to investigate the nature of the SWI/SNF complex following SMARCA4 mutation. We first evaluated the integrity of the SWI/SNF complex using human cancer cell lines that contain biallelic inactivating mutations in SMARCA4. We utilized SMARCC1 to purify the complex because it is present in all known variants of the SWI/SNF complex. Despite the absence of SMARCA4, SMARCC1 coprecipitated all 11 tested SWI/SNF subunits in the SMARCA4 mutant cell lines A549 and H1299 (Fig. 1A). Immunoprecipitation with antibodies to either SMARCA4 itself or control IgG did not yield SWI/SNF subunits, indicating that these associations are not due to nonspecific binding. Collectively, these findings establish that in SMARCA4 mutant cancer cell lines, other SWI/SNF subunits exist in a residual complex.
FIG 1.
Residual SWI/SNF complexes exist in SMARCA4 mutant cells and are essential for proliferation. (A) SMARCC1 associates with other SWI/SNF subunits in SMARCA4 mutant cancer cell lines. Immunoblots of SWI/SNF subunits before (input, 10%) and after precipitation (IP) with SMARCC1, SMARCA4, and IgG antibodies. Nuclear extracts used in these experiments are derived from the SMARCA4 mutant cell lines NCI-H1299 and A549. (B) SMARCB1 knockdown impairs proliferation of SMARCA4 mutant cancer cells. shRNA-mediated knockdown of SMARCB1 in SMARCA4 mutant NCI-H1299 and A549 cell lines is shown. Immunoblotting was used to determine the efficiency of knockdown. Cell proliferation was determined by cell count. Data are represented as means ± SEM from three biological replicates. (C) Reexpression of SMARCB1 rescues the growth defect. Shown is a colony formation assay following reexpression of a mouse derivative of SMARCB1 (mSMARCB1) that is not targeted by human SMARCB1 shRNAs. Images are from duplicate biological replicates. Immunoblotting was used to determine the efficiency of SMARCB1 knockdown.
Residual SWI/SNF complexes are essential in SMARCA4 mutant cell lines.
We next investigated whether residual SWI/SNF complexes were contributing to the growth of SMARCA4 mutant cancer cell lines. Using two different shRNAs, we knocked down the core SWI/SNF subunit, SMARCB1 (SNF5/INI1/BAF47), in SMARCA4 mutant cancer cell lines. We reasoned that since SMARCB1 is found in all variants of the SWI/SNF complex and is itself a tumor suppressor, reducing its levels would allow us to determine whether residual SWI/SNF complexes have a role in the viability of SMARCA4 mutant cancers. Using two different shRNAs targeting SMARCB1, we first tested the effects of SMARCB1 knockdown in two separate SMARCA4 mutant non-small-cell lung carcinoma cell lines. In both cases, reduced SMARCB1 levels markedly decreased proliferation, and the magnitude of effect correlated with the degree of knockdown (Fig. 1B). Reexpression of a mouse derivative of SMARCB1 not recognized by the human SMARCB1 shRNAs rescued the growth defects caused by SMARCB1 loss, indicating that the effect was on target (Fig. 1C). This finding establishes that SMARCB1 remains essential in the absence of SMARCA4.
SMARCA2 is differentially essential in SMARCA4 mutant cancer cell lines.
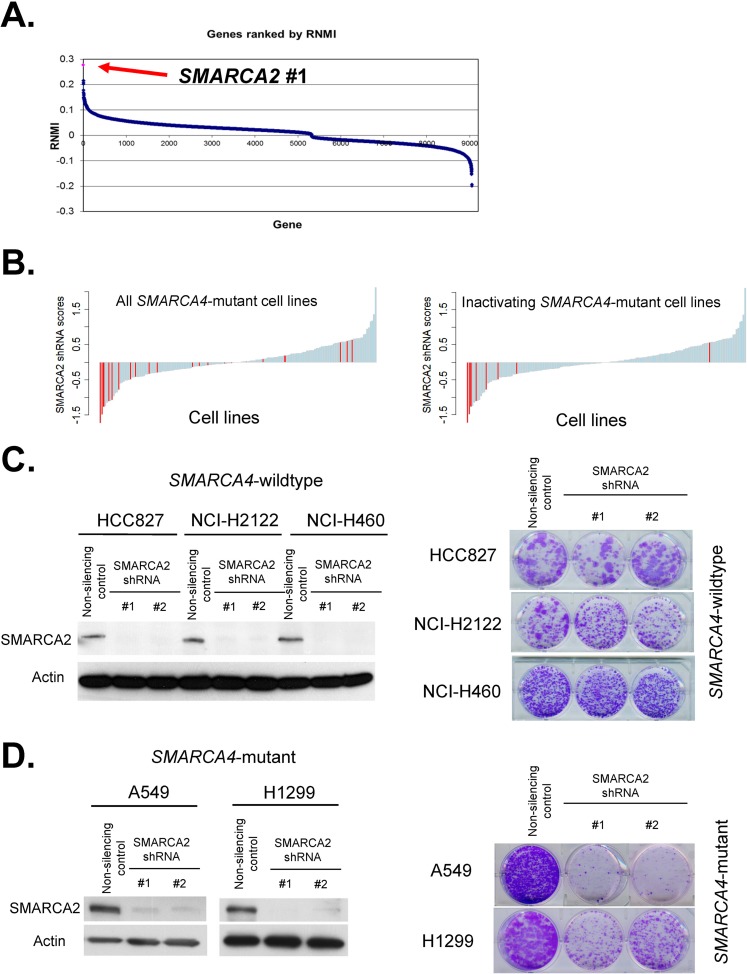
Independent of these experiments, we performed a large-scale screen to search for cancer-specific vulnerabilities created by SMARCA4 mutations. To do this, we used data from Project Achilles, a large-scale effort to identify essential genes in human cancer cell lines. Specifically, using data from the Cancer Cell Line Encyclopedia and prior publications (Table 1), we identified 21 cell lines in the Project Achilles screen that harbored mutations in SMARCA4 (http://www.broadinstitute.org/achilles) (26). We compared dependencies identified by screening 54,020 shRNAs targeting 11,194 genes in these 21 cancer cell lines to 144 SMARCA4 wild-type cell lines and identified SMARCA2 as the ninth most differentially essential gene in cancer cell lines containing mutations in SMARCA4 (P = 0.00069; false discovery rate [FDR], 0.69). As it is difficult to know whether individual missense mutations are driver or passenger changes, we next reanalyzed the data using only those cell lines that contained clear inactivating SMARCA4 mutations (frameshift, nonsense, and large focal deletions). When we compared dependencies in these 8 cell lines to the remaining 157 cell lines, SMARCA2 was the most differentially essential gene (P value = 7.366 × 10−06; FDR, <0.001) (Fig. 2A). Consistent with this, cell lines with inactivating SMARCA4 mutations were more sensitive to reduced levels of SMARCA2 than cell lines that have missense or noninactivating mutations (Fig. 2B). Using data from the Cancer Cell Line Encyclopedia, we next evaluated whether cell lines contained monoalleleic or biallelic inactivating mutations in SMARCA4. In six of the eight cell lines, mutations were biallelic (Table 1). In SK-MEL-5, only one allele has been found mutated, but this cell line has previously been shown to lack SMARCA4 expression, suggesting complete loss of function (29). In HEC1A, only one allele was mutated, and this cell line has previously been shown to express the SMARCA4 protein (30). Notably, HEC1A was the sole cell line with an inactivating SMARCA4 mutation that did not score as sensitive to SMARCA2 depletion. The SMARCA4 mutant cell lines used in these analyses were from various lineages (lung, ovary, endometrium, and skin), suggesting the dependency upon SMARCA2 is not a lineage-specific effect but occurs broadly in SMARCA4 mutant cancers. To verify the finding that SMARCA4 mutant cell lines were dependent upon SMARCA2 expression, we used independently derived shRNAs to suppress SMARCA2 levels in three SMARCA4 wild-type cell lines and two SMARCA4 mutant cell lines. While reduced SMARCA2 levels led to modest/no effects on the proliferation of SMARCA4 wild-type cell lines, SMARCA4 mutant cell lines were exquisitely sensitive to reduced SMARCA2 levels, as detected by colony formation assays (Fig. 2C and D). Despite reduced proliferation, we detected no significant change in apoptosis, senescence, or cell cycle (not shown), suggesting that combined loss of the ATPase subunits causes defects in multiple phases of the cell cycle. Indeed, the SWI/SNF complex has previously been shown to have functions during all four cell cycle phases (31–35).
FIG 2.
SMARCA2 is differentially essential in SMARCA4 mutant cancer cells. (A) Achilles shRNA screen identifies SMARCA2 as the number one vulnerability in SMARCA4-inactivated cancer cell lines. Gene scores from comparison of inactivating SMARCA4 mutant cancer cell lines to SMARCA4 wild-type cell lines are shown. (B) Cell lines with inactivating mutations in SMARCA4 are most sensitive to reduced levels of SMARCA2. Waterfall plot of Achilles SMARCA2 shRNA scores. Cell lines containing all SMARCA4 mutations are displayed in the left panel, and cell lines with inactivating SMARCA4 mutations (nonsense, frameshift, and large focal deletions) are displayed in the right panel. Red columns indicate SMARCA4 mutant cancer cell lines. (C) Reduced levels of SMARCA2 do not affect the growth of SMARCA4 wild-type cell lines. SMARCA2 knockdown in SMARCA4 wild-type cell lines HCC827, NCI-H2122, and H460. Immunoblotting is used to determine the efficiency of knockdown. Cell proliferation is evaluated using colony formation assays. Representative images are shown from one of two biological replicates for each cell line. (D) Reduced levels of SMARCA2 impair the growth of SMARCA4 mutant cell lines. SMARCA2 knockdown in SMARCA4 mutants NCI-H1299 and A549 leads to decreased proliferation. Immunoblotting is used to determine efficiency of knockdown. Cell proliferation is evaluated using colony formation assays. Representative images are shown from one of two biological replicates for each cell line.
SMARCA2 is largely dispensable for assembly of the SWI/SNF complex in SMARCA4 mutant cells.
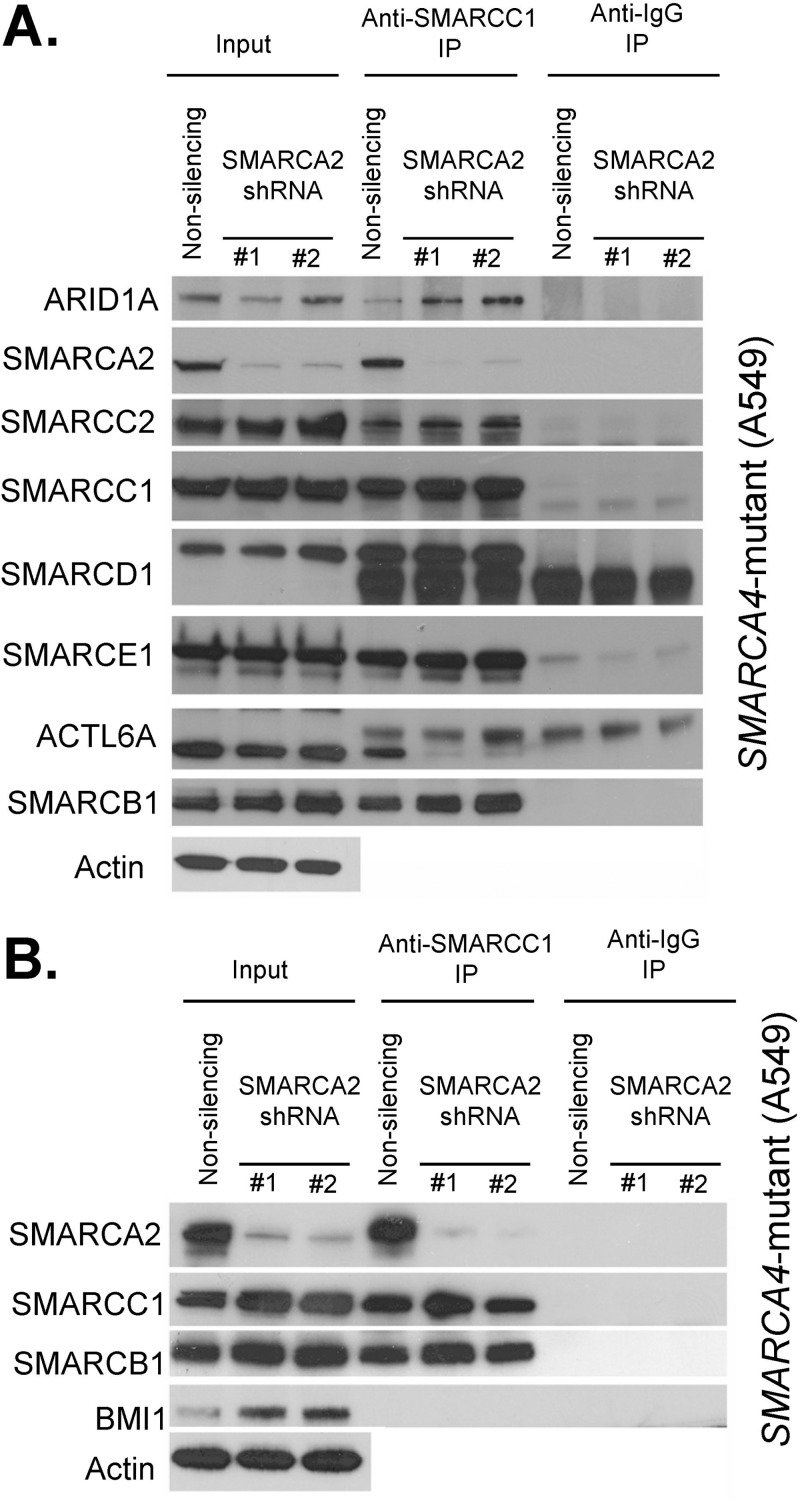
To gain insight into the specific vulnerability for SMARCA2 in SMARCA4 mutant cells, we next investigated whether knockdown of SMARCA2 affected the integrity of the residual SWI/SNF complex. Using coimmunoprecipitation of SMARCC1, we evaluated complex assembly following depletion of SMARCA2 in the A549 SMARCA4 mutant lung cancer cell line. Reduced levels of SMARCA2 had virtually no effect on the association between SMARCC1 and other SWI/SNF subunits, with a single exception (Fig. 3A). ACTL6A (BAF53) is a subunit of the SWI/SNF complex previously shown to bind directly to SMARCA4 (36). Following suppression of SMARCA2, ACTL6A no longer associated with SMARCC1. As a negative control to evaluate the specificity of these interactions, we evaluated BMI1, a subunit of the Polycomb chromatin complex not known to interact with the SWI/SNF complex, and found no association with SMARCC1 (Fig. 3B).
FIG 3.
SWI/SNF complex assembles in the absence of SMARCA2 and SMARCA4. (A) SMARCC1 associates with other SWI/SNF subunits following SMARCA2 knockdown in SMARCA4 mutant cells. Immunoblots of SWI/SNF subunits before (input, 10%) and after precipitation (IP) with SMARCC1 and IgG antibodies are shown. Nuclear extracts used in these experiments are derived from the SMARCA4 mutant cell line A549 treated with SMARCA2 shRNAs or nonsilencing control shRNAs. A representative image from duplicate biological replicates is shown. (B) SMARCC1 does not associate with the Polycomb-group protein BMI1. Immunoblots of inputs (input, 5%) and SMARCC1 and IgG precipitates (IP) following SMARCA2 knockdown in the SMARCA4 mutant cell line A549 are shown.
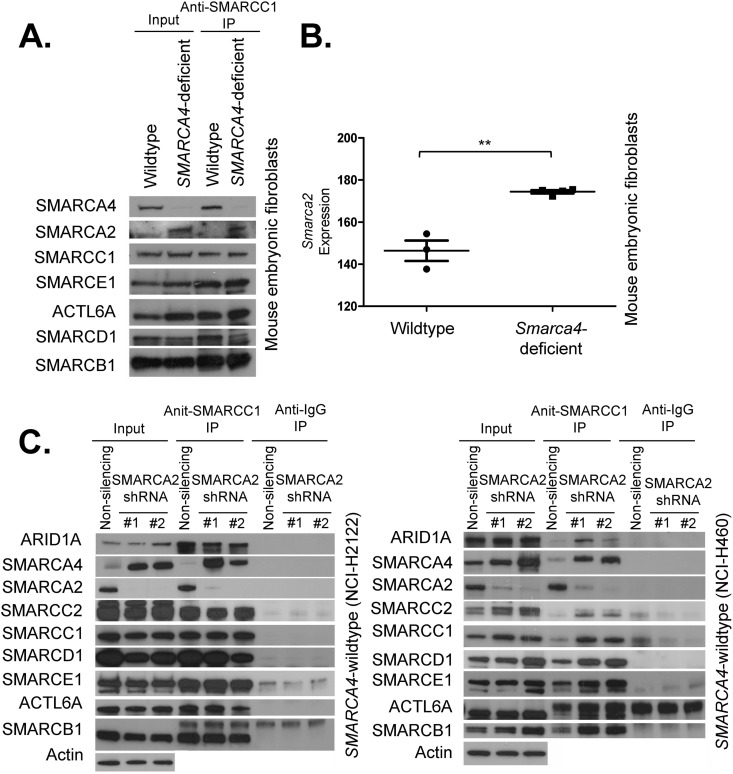
We next used genetically engineered Smarca4 conditional murine embryonic fibroblasts to evaluate the consequences of Smarca4 deletion upon expression of Smarca2 and incorporation of SMARCA2 into residual SWI/SNF complexes in primary cells. While the expression of other core SWI/SNF components was not affected by Smarca4 inactivation, SMARCA2 levels increased following Smarca4 inactivation, and SMARCA2 was incorporated in larger amounts into SWI/SNF complexes (Fig. 4A). This was due, at least in part, to increased transcription of the Smarca2 gene (P = 0.029) (Fig. 4B). Conversely, in SMARCA4 wild-type cells, knockdown of SMARCA2 led to a reciprocal effect of increased expression of SMARCA4, as well as increased assembly of SMARCA4 into the SWI/SNF complex (Fig. 4C). Collectively, these results indicate reciprocal expression and assembly of SMARCA2 and SMARCA4 into SWI/SNF complexes.
FIG 4.
Reciprocal compensatory functions for SMARCA2 and SMARCA4. (A) Smarca4 inactivation in primary cells leads to specific upregulation of Smarca2 and increased association of SMARCA2 with other SWI/SNF subunits. An immunoblot of SWI/SNF subunits before (input, 10%) and after precipitation (IP) with SMARCC1 is shown. Nuclear extracts used in these experiments are derived from wild-type and Smarca4-deficient mouse embryonic fibroblasts. A representative image from duplicate biological replicates is shown. (B) Smarca2 transcript levels are elevated following Smarca4 inactivation. A plot of Smarca2 transcript expression levels in wild-type and Smarca4-deficient mouse embryonic fibroblasts is shown. **, P = 0.029. (C) SMARCA2 knockdown leads to SMARCA4 upregulation and increased association of SMARCA4 with other SWI/SNF subunits. Immunoblots of SWI/SNF subunits before (input, 10%) and after precipitation (IP) with SMARCC1 and IgG antibodies. Nuclear extracts used in these experiments are derived from the SMARCA4 wild-type cell lines NCI-H2122 and H460 treated with SMARCA2 shRNAs or nonsilencing control shRNAs. Representative images from duplicate biological replicates are shown.
DISCUSSION
One-fifth of all human cancers have mutations in genes encoding subunits of the SWI/SNF chromatin remodeling complex (2). As SMARCA4 is recurrently mutated in several cancer types, the identification of cancer-specific vulnerabilities created by mutations in the SMARCA4 subunit carries substantial relevance to human disease. Given that many SMARCA4 mutations are clearly inactivating, a key question is whether mutations in SMARCA4 lead to a complete loss of function of the SWI/SNF complex or whether the remaining SWI/SNF subunits assemble into residual complexes that can serve as therapeutic targets in SMARCA4 mutant cancers. Here, we demonstrate that SMARCB1, a core SWI/SNF subunit found in all variant SWI/SNF complexes, is essential in SMARCA4 mutant cancer cells, suggesting important roles for residual complexes in the tumorigenesis of SMARCA4 mutant cancers. We further demonstrate a specific dependency upon the mutually exclusive SWI/SNF subunit SMARCA2 in these cancers, implicating SMARCA2-containing residual complexes as essential for tumorigenesis following SMARCA4 loss. As we were preparing our manuscript, a study that tested the hypothesis that SMARCA2 takes over for SMARCA4 in cancer cells was published, and it similarly concluded that SMARCA2 was specifically essential in SMARCA4 mutant cancer cells (37). Notably, SMARCB1 depletion did not score as differentially essential for SMARCA4 mutant cell lines in our vulnerability screen, consistent with its essential role in early mouse development and its essential role in most normal lineages (25, 38–40). Collectively, our work identifies SMARCA2-containing residual complexes as a specific vulnerability in SMARCA4 mutant cancers and implicates SMARCA2 as a therapeutic target in these cancers. In future experiments, it will be of interest to investigate the mechanism underlying the oncogenic drive. Is it loss of some aspect of SMARCA4 function and SMARCA2 simply rescues the loss of viability that would otherwise be caused by SMARCA4 loss, or does the expression of SMARCA2 bring neomorphic protransformation activity to the complex?
Our studies also indicate a reciprocal assembly of SMARCA2 into SWI/SNF complexes following genetic inactivation of SMARCA4, providing insight into the mechanisms driving tumorigenesis. Previous studies have suggested compensatory roles for SMARCA2 and SMARCA4 (23, 24, 41, 42). Our studies similarly implicate a reciprocal regulation of SMARCA2 and SMARCA4 that contributes to tumorigenesis. However, we also demonstrate distinct roles for SMARCA2 and SMARCA4 during oncogenic transformation by showing essential roles for SMARCA2 in SMARCA4 mutant cancers. Further highlighting functional differences between SMARCA4 and SMARCA2, homozygous inactivation of SMARCA4 is embryonic lethal, whereas SMARCA2-deficient mice are viable (11, 23). SMARCA2 and SMARCA4 are also expressed differentially during development and have distinct chromatin binding profiles and differential effects on gene regulation (21, 43). SMARCA4 also associates with transcription factors that are distinct from those that associate with SMARCA2 (43). Unlike SMARCA4, missense mutations of SMARCA2 that have unknown functional impact, whether gain of function, loss of function, or dominant negative, occur in Nicolaides-Baraitser syndrome (44–46). Lastly, during osteoblast differentiation, SMARCA2 and SMARCA4 have been shown to serve opposing functions with respect to differentiation (47). Collectively, our work demonstrates reciprocal regulation and assembly of SMARCA4 and SMARCA2 and demonstrates partial redundancy such that SMARCA2 becomes a specific vulnerability in SMARCA4 mutant cancers.
SMARCA4 is frequently mutated in cancer. Inactivating SMARCA2 mutations are rare in cancer, but cell lines and primary cancers have been identified that contain little or no SMARCA2 (48, 49). It is unclear whether there are low but essential levels of SMARCA2 in these cases, contributing to vulnerability, or whether in some cases cancers can adapt to the complete absence of both SMARCA4 and SMARCA2. Future studies are needed to determine whether functional residual complexes exist in these cancers and whether they contribute to tumorigenesis. Further work is also needed to uncover the mechanisms driving tumorigenesis in cells that may have become resistant to SMARCA2 loss despite the absence of SMARCA4.
Therapeutic targets for SMARCA4 mutant cancers have remained elusive. Our studies suggest that therapeutic inhibition of SMARCA2-containing residual complexes is a promising approach for the treatment of SMARCA4 mutant cancers. Notably, SMARCA2-deficient mice are viable, suggesting that SMARCA2 generally is not required for cellular proliferation. SMARCA2 harbors at least two potentially targetable domains, an enzymatic ATPase domain and a bromodomain. Bromodomains have recently emerged as particularly good targets for anticancer therapy (50). Collectively, these studies identify SMARCA2 as a viable therapeutic target in SMARCA4 mutant cancers.
ACKNOWLEDGMENTS
We thank J. Shapiro for providing us with control SMARCA4 wild-type cell lines. We thank Andrew J. Aguirre for helpful discussion regarding the analysis of Achilles data.
B.G.W. was supported by a Childhood Cancer Research Grant from the Pablove Foundation. X.W. was supported by a postdoctoral fellowship from the David Abraham Foundation and Rally Foundation. This work was supported by R01CA172152 (C.W.M.R.) and R01CA113794 (C.W.M.R.), as well as by a U01 NCI Mouse Models of Cancer Consortium Award (C.W.M.R.). The Garrett B. Smith Foundation, Miles for Mary, and the Cure AT/RT Now foundation (C.W.M.R.) provided additional support.
We declare no conflicts of interest.
Footnotes
Published ahead of print 13 January 2014
REFERENCES
- 1.Wilson BG, Roberts CW. 2011. SWI/SNF nucleosome remodellers and cancer. Nat. Rev. Cancer 11:481–492. 10.1038/nrc3068 [DOI] [PubMed] [Google Scholar]
- 2.Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J, Crabtree GR. 2013. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat. Genet. 45:592–601. 10.1038/ng.2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shain AH, Pollack JR. 2013. The spectrum of SWI/SNF mutations, ubiquitous in human cancers. PLoS One 8:e55119. 10.1371/journal.pone.0055119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weissman B, Knudsen KE. 2009. Hijacking the chromatin remodeling machinery: impact of SWI/SNF perturbations in cancer. Cancer Res. 69:8223–8230. 10.1158/0008-5472.CAN-09-2166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reisman D, Glaros S, Thompson EA. 2009. The SWI/SNF complex and cancer. Oncogene 28:1653–1668. 10.1038/onc.2009.4 [DOI] [PubMed] [Google Scholar]
- 6.Love C, Sun Z, Jima D, Li G, Zhang J, Miles R, Richards KL, Dunphy CH, Choi WW, Srivastava G, Lugar PL, Rizzieri DA, Lagoo AS, Bernal-Mizrachi L, Mann KP, Flowers CR, Naresh KN, Evens AM, Chadburn A, Gordon LI, Czader MB, Gill JI, Hsi ED, Greenough A, Moffitt AB, McKinney M, Banerjee A, Grubor V, Levy S, Dunson DB, Dave SS. 2012. The genetic landscape of mutations in Burkitt lymphoma. Nat. Genet. 44:1321–1325. 10.1038/ng.2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C, Dicara D, Ramos AH, Lawrence MS, Cibulskis K, Sivachenko A, Voet D, Saksena G, Stransky N, Onofrio RC, Winckler W, Ardlie K, Wagle N, Wargo J, Chong K, Morton DL, Stemke-Hale K, Chen G, Noble M, Meyerson M, Ladbury JE, Davies MA, Gershenwald JE, Wagner SN, Hoon DS, Schadendorf D, Lander ES, Gabriel SB, Getz G, Garraway LA, Chin L. 2012. A landscape of driver mutations in melanoma. Cell 150:251–263. 10.1016/j.cell.2012.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parsons DW, Li M, Zhang X, Jones S, Leary RJ, Lin JC, Boca SM, Carter H, Samayoa J, Bettegowda C, Gallia GL, Jallo GI, Binder ZA, Nikolsky Y, Hartigan J, Smith DR, Gerhard DS, Fults DW, Vandenberg S, Berger MS, Marie SK, Shinjo SM, Clara C, Phillips PC, Minturn JE, Biegel JA, Judkins AR, Resnick AC, Storm PB, Curran T, He Y, Rasheed BA, Friedman HS, Keir ST, McLendon R, Northcott PA, Taylor MD, Burger PC, Riggins GJ, Karchin R, Parmigiani G, Bigner DD, Yan H, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE. 2011. The genetic landscape of the childhood cancer medulloblastoma. Science 331:435–439. 10.1126/science.1198056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Medina PP, Romero OA, Kohno T, Montuenga LM, Pio R, Yokota J, Sanchez-Cespedes M. 2008. Frequent BRG1/SMARCA4-inactivating mutations in human lung cancer cell lines. Hum. Mutat. 29:617–622. 10.1002/humu.20730 [DOI] [PubMed] [Google Scholar]
- 10.Shain AH, Giacomini CP, Matsukuma K, Karikari CA, Bashyam MD, Hidalgo M, Maitra A, Pollack JR. 2012. Convergent structural alterations define SWItch/Sucrose NonFermentable (SWI/SNF) chromatin remodeler as a central tumor suppressive complex in pancreatic cancer. Proc. Natl. Acad. Sci. U. S. A. 109:E252–E259. 10.1073/pnas.1114817109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, Randazzo F, Metzger D, Chambon P, Crabtree G, Magnuson T. 2000. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell 6:1287–1295. 10.1016/S1097-2765(00)00127-1 [DOI] [PubMed] [Google Scholar]
- 12.Bultman SJ, Herschkowitz JI, Godfrey V, Gebuhr TC, Yaniv M, Perou CM, Magnuson T. 2008. Characterization of mammary tumors from Brg1 heterozygous mice. Oncogene 27:460–468. 10.1038/sj.onc.1210664 [DOI] [PubMed] [Google Scholar]
- 13.Glaros S, Cirrincione GM, Palanca A, Metzger D, Reisman D. 2008. Targeted knockout of BRG1 potentiates lung cancer development. Cancer Res. 68:3689–3696. 10.1158/0008-5472.CAN-07-6652 [DOI] [PubMed] [Google Scholar]
- 14.Khavari PA, Peterson CL, Tamkun JW, Mendel DB, Crabtree GR. 1993. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature 366:170–174. 10.1038/366170a0 [DOI] [PubMed] [Google Scholar]
- 15.Muchardt C, Yaniv M. 1993. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 12:4279–4290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiba H, Muramatsu M, Nomoto A, Kato H. 1994. Two human homologues of Saccharomyces cerevisiae SWI2/SNF2 and Drosophila brahma are transcriptional coactivators cooperating with the estrogen receptor and the retinoic acid receptor. Nucleic Acids Res. 22:1815–1820. 10.1093/nar/22.10.1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang W, Cote J, Xue Y, Zhou S, Khavari PA, Biggar SR, Muchardt C, Kalpana GV, Goff SP, Yaniv M, Workman JL, Crabtree GR. 1996. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 15:5370–5382 [PMC free article] [PubMed] [Google Scholar]
- 18.de la Serna IL, Ohkawa Y, Imbalzano AN. 2006. Chromatin remodelling in mammalian differentiation: lessons from ATP-dependent remodellers. Nat. Rev. Genet. 7:461–473. 10.1038/nrg1882 [DOI] [PubMed] [Google Scholar]
- 19.Yoo AS, Crabtree GR. 2009. ATP-dependent chromatin remodeling in neural development. Curr. Opin. Neurobiol. 19:120–126. 10.1016/j.conb.2009.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohrmann L, Verrijzer CP. 2005. Composition and functional specificity of SWI2/SNF2 class chromatin remodeling complexes. Biochim. Biophys. Acta 1681:59–73. 10.1016/j.bbaexp.2004.10.005 [DOI] [PubMed] [Google Scholar]
- 21.Kaeser MD, Aslanian A, Dong MQ, Yates JR, III, Emerson BM. 2008. BRD7, a novel PBAF-specific SWI/SNF subunit, is required for target gene activation and repression in embryonic stem cells. J. Biol. Chem. 283:32254–32263. 10.1074/jbc.M806061200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reisman DN, Sciarrotta J, Bouldin TW, Weissman BE, Funkhouser WK. 2005. The expression of the SWI/SNF ATPase subunits BRG1 and BRM in normal human tissues. Appl. Immunohistochem. Mol. Morphol. 13:66–74. 10.1097/00129039-200503000-00011 [DOI] [PubMed] [Google Scholar]
- 23.Reyes JC, Barra J, Muchardt C, Camus A, Babinet C, Yaniv M. 1998. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2alpha). EMBO J. 17:6979–6991. 10.1093/emboj/17.23.6979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, Sansam CG, Thom CS, Metzger D, Evans JA, Nguyen PT, Roberts CW. 2009. Oncogenesis caused by loss of the SNF5 tumor suppressor is dependent on activity of BRG1, the ATPase of the SWI/SNF chromatin remodeling complex. Cancer Res. 69:8094–8101. 10.1158/0008-5472.CAN-09-0733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Isakoff MS, Sansam CG, Tamayo P, Subramanian A, Evans JA, Fillmore CM, Wang X, Biegel JA, Pomeroy SL, Mesirov JP, Roberts CW. 2005. Inactivation of the Snf5 tumor suppressor stimulates cell cycle progression and cooperates with p53 loss in oncogenic transformation. Proc. Natl. Acad. Sci. U. S. A. 102:17745–17750. 10.1073/pnas.0509014102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheung HW, Cowley GS, Weir BA, Boehm JS, Rusin S, Scott JA, East A, Ali LD, Lizotte PH, Wong TC, Jiang G, Hsiao J, Mermel CH, Getz G, Barretina J, Gopal S, Tamayo P, Gould J, Tsherniak A, Stransky N, Luo B, Ren Y, Drapkin R, Bhatia SN, Mesirov JP, Garraway LA, Meyerson M, Lander ES, Root DE, Hahn WC. 2011. Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proc. Natl. Acad. Sci. U. S. A. 108:12372–12377. 10.1073/pnas.1109363108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shao DD, Tsherniak A, Gopal S, Weir BA, Tamayo P, Stransky N, Schumacher SE, Zack TI, Beroukhim R, Garraway LA, Margolin AA, Root DE, Hahn WC, Mesirov JP. 2012. ATARiS: computational quantification of gene suppression phenotypes from multi-sample RNAi screens. Genome Res. 23:665–678. 10.1101/gr.143586.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tolstorukov MY, Sansam CG, Lu P, Koellhoffer EC, Helming KC, Alver BH, Tillman EJ, Evans JA, Wilson BG, Park PJ, Roberts CW. 2013. Swi/Snf chromatin remodeling/tumor suppressor complex establishes nucleosome occupancy at target promoters. Proc. Natl. Acad. Sci. U. S. A. 110:10165–10170. 10.1073/pnas.1302209110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keenen B, Qi H, Saladi SV, Yeung M, de la Serna IL. 2010. Heterogeneous SWI/SNF chromatin remodeling complexes promote expression of microphthalmia-associated transcription factor target genes in melanoma. Oncogene 29:81–92. 10.1038/onc.2009.304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guan B, Wang TL, Shih IM. 2011. ARID1A, a factor that promotes formation of SWI/SNF-mediated chromatin remodeling, is a tumor suppressor in gynecologic cancers. Cancer Res. 71:6718–6727. 10.1158/0008-5472.CAN-11-1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dykhuizen EC, Hargreaves DC, Miller EL, Cui K, Korshunov A, Kool M, Pfister S, Cho YJ, Zhao K, Crabtree GR. 2013. BAF complexes facilitate decatenation of DNA by topoisomerase IIalpha. Nature 497:624–627. 10.1038/nature12146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Versteege I, Medjkane S, Rouillard D, Delattre O. 2002. A key role of the hSNF5/INI1 tumour suppressor in the control of the G1-S transition of the cell cycle. Oncogene 21:6403–6412. 10.1038/sj.onc.1205841 [DOI] [PubMed] [Google Scholar]
- 33.Trouche D, Le Chalony C, Muchardt C, Yaniv M, Kouzarides T. 1997. RB and hbrm cooperate to repress the activation functions of E2F1. Proc. Natl. Acad. Sci. U. S. A. 94:11268–11273. 10.1073/pnas.94.21.11268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xue Y, Canman JC, Lee CS, Nie Z, Yang D, Moreno GT, Young MK, Salmon ED, Wang W. 2000. The human SWI/SNF-B chromatin-remodeling complex is related to yeast rsc and localizes at kinetochores of mitotic chromosomes. Proc. Natl. Acad. Sci. U. S. A. 97:13015–13020. 10.1073/pnas.240208597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sif S, Stukenberg PT, Kirschner MW, Kingston RE. 1998. Mitotic inactivation of a human SWI/SNF chromatin remodeling complex. Genes Dev. 12:2842–2851. 10.1101/gad.12.18.2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao K, Wang W, Rando OJ, Xue Y, Swiderek K, Kuo A, Crabtree GR. 1998. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell 95:625–636. 10.1016/S0092-8674(00)81633-5 [DOI] [PubMed] [Google Scholar]
- 37.Oike T, Ogiwara H, Tominaga Y, Ito K, Ando O, Tsuta K, Mizukami T, Shimada Y, Isomura H, Komachi M, Furuta K, Watanabe SI, Nakano T, Yokota J, Kohno T. 2013. A synthetic lethality-based strategy to treat cancers harboring a genetic deficiency in the chromatin remodeling factor BRG1. Cancer Res. 73:5508–5518. 10.1158/0008-5472.CAN-12-4593 [DOI] [PubMed] [Google Scholar]
- 38.Gresh L, Bourachot B, Reimann A, Guigas B, Fiette L, Garbay S, Muchardt C, Hue L, Pontoglio M, Yaniv M, Klochendler-Yeivin A. 2005. The SWI/SNF chromatin-remodeling complex subunit SNF5 is essential for hepatocyte differentiation. EMBO J. 24:3313–3324. 10.1038/sj.emboj.7600802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roberts CW, Leroux MM, Fleming MD, Orkin SH. 2002. Highly penetrant, rapid tumorigenesis through conditional inversion of the tumor suppressor gene Snf5. Cancer Cell 2:415–425. 10.1016/S1535-6108(02)00185-X [DOI] [PubMed] [Google Scholar]
- 40.Wang X, Werneck MB, Wilson BG, Kim HJ, Kluk MJ, Thom CS, Wischhusen JW, Evans JA, Jesneck JL, Nguyen P, Sansam CG, Cantor H, Roberts CW. 2011. TCR-dependent transformation of mature memory phenotype T cells in mice. J. Clin. Investig. 121:3834–3845. 10.1172/JCI37210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Willis MS, Homeister JW, Rosson GB, Annayev Y, Holley D, Holly SP, Madden VJ, Godfrey V, Parise LV, Bultman SJ. 2012. Functional redundancy of SWI/SNF catalytic subunits in maintaining vascular endothelial cells in the adult heart. Circ. Res. 111:e111–e122. 10.1161/CIRCRESAHA.112.265587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strobeck MW, Reisman DN, Gunawardena RW, Betz BL, Angus SP, Knudsen KE, Kowalik TF, Weissman BE, Knudsen ES. 2002. Compensation of BRG-1 function by Brm: insight into the role of the core SWI-SNF subunits in retinoblastoma tumor suppressor signaling. J. Biol. Chem. 277:4782–4789. 10.1074/jbc.M109532200 [DOI] [PubMed] [Google Scholar]
- 43.Kadam S, Emerson BM. 2003. Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol. Cell 11:377–389. 10.1016/S1097-2765(03)00034-0 [DOI] [PubMed] [Google Scholar]
- 44.Wieczorek D, Bogershausen N, Beleggia F, Steiner-Haldenstatt S, Pohl E, Li Y, Milz E, Martin M, Thiele H, Altmuller J, Alanay Y, Kayserili H, Klein-Hitpass L, Bohringer S, Wollstein A, Albrecht B, Boduroglu K, Caliebe A, Chrzanowska K, Cogulu O, Cristofoli F, Czeschik JC, Devriendt K, Dotti MT, Elcioglu N, Gener B, Goecke TO, Krajewska-Walasek M, Guillen-Navarro E, Hayek J, Houge G, Kilic E, Simsek-Kiper PO, Lopez-Gonzalez V, Kuechler A, Lyonnet S, Mari F, Marozza A, Mathieu Dramard M, Mikat B, Morin G, Morice-Picard F, Ozkinay F, Rauch A, Renieri A, Tinschert S, Utine GE, Vilain C, Vivarelli R, Zweier C, Nurnberg P, Rahmann S, Vermeesch J, Ludecke HJ, Zeschnigk M, Wollnik B. 2013. A comprehensive molecular study on Coffin-Siris and Nicolaides-Baraitser syndromes identifies a broad molecular and clinical spectrum converging on altered chromatin remodeling. Hum. Mol. Genet. 22:5121–5135. 10.1093/hmg/ddt366 [DOI] [PubMed] [Google Scholar]
- 45.Van Houdt JK, Nowakowska BA, Sousa SB, van Schaik BD, Seuntjens E, Avonce N, Sifrim A, Abdul-Rahman OA, van den Boogaard MJ, Bottani A, Castori M, Cormier-Daire V, Deardorff MA, Filges I, Fryer A, Fryns JP, Gana S, Garavelli L, Gillessen-Kaesbach G, Hall BD, Horn D, Huylebroeck D, Klapecki J, Krajewska-Walasek M, Kuechler A, Lines MA, Maas S, Macdermot KD, McKee S, Magee A, de Man SA, Moreau Y, Morice-Picard F, Obersztyn E, Pilch J, Rosser E, Shannon N, Stolte-Dijkstra I, Van Dijck P, Vilain C, Vogels A, Wakeling E, Wieczorek D, Wilson L, Zuffardi O, van Kampen AH, Devriendt K, Hennekam R, Vermeesch JR. 2012. Heterozygous missense mutations in SMARCA2 cause Nicolaides-Baraitser syndrome. Nat. Genet. 44:445–449. 10.1038/ng.1105 [DOI] [PubMed] [Google Scholar]
- 46.Wolff D, Endele S, Azzarello-Burri S, Hoyer J, Zweier M, Schanze I, Schmitt B, Rauch A, Reis A, Zweier C. 2012. In-frame deletion and missense mutations of the C-terminal helicase domain of SMARCA2 in three patients with Nicolaides-Baraitser syndrome. Mol. Syndromol. 2:237–244. 10.1159/000337323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Flowers S, Nagl NG, Jr, Beck GR, Jr, Moran E. 2009. Antagonistic roles for BRM and BRG1 SWI/SNF complexes in differentiation. J. Biol. Chem. 284:10067–10075. 10.1074/jbc.M808782200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reisman DN, Strobeck MW, Betz BL, Sciariotta J, Funkhouser W, Jr, Murchardt C, Yaniv M, Sherman LS, Knudsen ES, Weissman BE. 2002. Concomitant down-regulation of BRM and BRG1 in human tumor cell lines: differential effects on RB-mediated growth arrest vs CD44 expression. Oncogene 21:1196–1207. 10.1038/sj.onc.1205188 [DOI] [PubMed] [Google Scholar]
- 49.Yamamichi N, Inada K, Ichinose M, Yamamichi-Nishina M, Mizutani T, Watanabe H, Shiogama K, Fujishiro M, Okazaki T, Yahagi N, Haraguchi T, Fujita S, Tsutsumi Y, Omata M, Iba H. 2007. Frequent loss of Brm expression in gastric cancer correlates with histologic features and differentiation state. Cancer Res. 67:10727–10735. 10.1158/0008-5472.CAN-07-2601 [DOI] [PubMed] [Google Scholar]
- 50.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, Philpott M, Munro S, McKeown MR, Wang Y, Christie AL, West N, Cameron MJ, Schwartz B, Heightman TD, La Thangue N, French CA, Wiest O, Kung AL, Knapp S, Bradner JE. 2010. Selective inhibition of BET bromodomains. Nature 468:1067–1073. 10.1038/nature09504 [DOI] [PMC free article] [PubMed] [Google Scholar]