Abstract
Anaerobic gut fungi represent a distinct early-branching fungal phylum (Neocallimastigomycota) and reside in the rumen, hindgut, and feces of ruminant and nonruminant herbivores. The genome of an anaerobic fungal isolate, Orpinomyces sp. strain C1A, was sequenced using a combination of Illumina and PacBio single-molecule real-time (SMRT) technologies. The large genome (100.95 Mb, 16,347 genes) displayed extremely low G+C content (17.0%), large noncoding intergenic regions (73.1%), proliferation of microsatellite repeats (4.9%), and multiple gene duplications. Comparative genomic analysis identified multiple genes and pathways that are absent in Dikarya genomes but present in early-branching fungal lineages and/or nonfungal Opisthokonta. These included genes for posttranslational fucosylation, the production of specific intramembrane proteases and extracellular protease inhibitors, the formation of a complete axoneme and intraflagellar trafficking machinery, and a near-complete focal adhesion machinery. Analysis of the lignocellulolytic machinery in the C1A genome revealed an extremely rich repertoire, with evidence of horizontal gene acquisition from multiple bacterial lineages. Experimental analysis indicated that strain C1A is a remarkable biomass degrader, capable of simultaneous saccharification and fermentation of the cellulosic and hemicellulosic fractions in multiple untreated grasses and crop residues examined, with the process significantly enhanced by mild pretreatments. This capability, acquired during its separate evolutionary trajectory in the rumen, along with its resilience and invasiveness compared to prokaryotic anaerobes, renders anaerobic fungi promising agents for consolidated bioprocessing schemes in biofuels production.
INTRODUCTION
Members of the anaerobic gut fungi were originally discovered in sheep (1) but have subsequently been observed in the rumen, hindgut, and feces of ruminant and nonruminant herbivorous mammals and reptilian herbivores. The observation of flagellated zoospores of anaerobic fungi was reported as early as 1910 (2). However, the accidental discovery and subsequent proof that these flagellated zoospores were actually spores of a new fungal lineage rather than ciliated protozoa came relatively late (1). Anaerobic gut fungi belong to the phylum Neocallimastigomycota, an early-branching fungal lineage, for which no current genome analysis has yet been reported. With the exception of the Microsporidia, few genomes belonging to non-Dikarya fungal lineages have been sequenced and analyzed (3, 4). Therefore, analysis of a Neocallimastigomycota genome and genomic analytic comparison to early-branching and Dikarya fungal genomes could identify salient characteristics associated with fungal evolution and diversification.
In addition to their distinct phylogenetic position, anaerobic fungi appear to be habitat restricted and are the only known fungal group that lives within the rumen and gut of herbivores (5). This evolutionary trajectory in a distinct habitat resulted in multiple metabolic and structural adaptations. For example, members of the Neocallimastigomycota have adapted a strict anaerobic lifestyle. Similar to other anaerobic eukaryotes (e.g., Trichomonas vaginalis [6, 7]), their mitochondria have undergone a reductive evolution process to a hydrogenosome, an organelle whose main function is ATP production via substrate-level phosphorylation and hydrogen production (6, 8, 9). Anaerobic fungi also reproduce asexually via the release of motile flagellated zoospores from zoosporangia that develop during rhizoidal fungal growth (10, 11). Finally, anaerobic fungi are highly fibrolytic microorganisms, producing a wide array of cellulolytic, hemicellulolytic, glycolytic, and proteolytic enzymes (5, 12–15). It has been established that in anaerobic gut habitats, these organisms play a role akin to their aerobic counterparts in soils and streams. By attaching themselves to plant materials, they colonize and excrete extracellular enzymes that mobilize the structural plant polymers to be available to other microbes.
Therefore, analysis of Neocallimastigomycota genomes could not only lead to better understanding of the genomic features and metabolic characteristics of an early-branching fungal lineage but also lead to the identification of metabolic, physiological, and genome-wide adaptations that enabled the survival and establishment of Neocallimastigomycota as core members of the highly eutrophic, prokaryote-dominated herbivorous rumen and gut. Here, we report on the sequencing and analysis of the draft genome and transcriptome of the anaerobic fungal isolate Orpinomyces sp. strain C1A (henceforth C1A). We identified multiple unique features within the genome and reason that these genomic features are a reflection of two important factors: its placement within a phylogenetically distinct early-branching phylum in the Mycota and its adaptation to the animal rumen gut during its separate evolutionary trajectory from the Mycota. We further demonstrate that one of these evolutionary adaptations, the presence of a remarkably efficient lignocellulolytic machinery coupled to anaerobic fermentative metabolism of hexose and pentose monomers, renders this microorganism an extremely promising agent for lignocellulolytic conversion in consolidated biological processing (CBP) schemes for biofuel production.
MATERIALS AND METHODS
Culturing, DNA sequencing, and genome assembly. (i) Culturing.
Strain C1A was isolated from the feces of an Angus steer on a cellobiose-switchgrass medium using previously described protocols (16). Orpinomyces strain C1A was grown in an anaerobic, rumen fluid-free basal medium that was reduced by cysteine-sulfide and dispensed under a stream of 100% CO2 as previously described (17). Cellobiose (3.75 g/liter) was used as the substrate. C1A cultures were scaled up for nucleic acid extraction in 1-liter batches prepared in 2-liter Schott bottles equipped with the stoppered top of a Balch tube to maintain strict anaerobic conditions during fungal growth. Culturing was conducted using the techniques described by Bryant and modified by Balch and Wolfe (18, 19). After autoclaving, the Schott bottles were cooled to room temperature and the gas phase was replaced by vacuuming and repressurization with 100% CO2 (19). The medium was then amended with penicillin, streptomycin, and chloramphenicol from an anaerobic stock solution in order to provide final concentrations of 50 μg/ml, 20 μg/ml, and 50 μg/ml of each antibiotic, respectively. The medium was then prewarmed at 39°C for approximately 3 to 4 h and inoculated with 50 ml of an actively growing culture of Orpinomyces strain C1A. The cultures were incubated at 39°C for approximately 3 to 4 days, and the fungal cells were harvested during late log phase by centrifugation at 10,000 rpm for 30 min.
(ii) DNA extraction and sequencing.
(a) Illumina sequencing. High-molecular-weight genomic DNA was extracted using a modified cetyltrimethylammonium bromide (CTAB) method for isolation of nucleic acids in anaerobic fungi (20). Four micrograms of high-molecular-weight DNA was used to generate libraries for Illumina Sequence by Synthesis (Illumina-SBS) genome sequencing (21) using the standard Illumina TruSeq DNA protocol (http://genome.med.harvard.edu/documents/illumina/TruSeq_DNA_SamplePrep_Guide_15005180_A.pdf). Post-adaptor ligation size-selected fragments used for flow-cell cluster generation had a mean size of 293 bp as reported by the Agilent 3200 Bioanalyzer. Illumina sequencing was conducted using the services of a commercial provider (Ambrys Genetics, Aliso Viejo, CA, USA) on a HiSeq 2000 sequencing platform using 100-bp paired-end chemistry. Illumina sequencing yielded 29.2 Gb in 146,385,792 quality-filtered paired-end reads (see Table S1 in the supplemental material).
(b) PacBio sequencing. DNA used for Pacific Biosciences (PacBio) single-molecule real-time (SMRT) sequencing (22) was isolated using the Epicentre plant DNA extraction kit (Epicentre Corp., Madison, WI, USA) according to the manufacturer's specification. Selected inserts of 5- to 10-kb read size were prepared from 10 μg of extracted high-molecular-weight DNA by ligation to the SMRTbell sequencing adaptor. SMRT sequencing was conducted using the services of a commercial provider (Expression Analysis, Durham, NC, USA) on a PacBio RS sequencing platform using the second-generation C2 sequencing chemistry with eight zero-mode wavelength (ZMW) SMRT cells. PacBio sequencing yielded 984.8 Mb of quality-filtered data in 463,832 raw long reads (see Table S1 in the supplemental material), with an average read length of 2,124 bp. An additional 26.9 Mb of DNA sequence data was harbored in 16,949 reads (average read length of 1,586 bp) that reached a circular consensus (CCS) during sequencing and was used for long read error correction.
(iii) Genome assembly.
All computational assemblies were conducted using the SGI UV 1000 cache coherent nonuniform memory architecture (cc-NUMA) high-performance computing system Blacklight. Blacklight is an Extreme Science and Engineering Discovery Environment (XSEDE) community-shared computational resource dedicated for high-memory-footprint jobs such as de novo assembly. Blacklight is housed at the Pittsburgh Supercomputing Center (http://www.psc.edu/).
We initially attempted to utilize Illumina paired-end sequencing as the sole mechanism for C1A genome sequencing. Illumina quality-filtered reads were assembled with Velvet 1.1.07 (23), using a kmer value of 63 and a minimum coverage cutoff of 7. The resulting assembly was highly fragmented (see Table S2 in the supplemental material), with an extremely large number of contigs in the final assembly (82,325 contigs), a large proportion of the final assembly (32.4%) harbored in extremely short contigs (300 to 900 bp), and a low N50 (1,666 bp).
Therefore, we sought to improve the assembly by using a hybrid SMRT-Illumina strategy that leverages short-read high-accuracy data formed from Illumina sequencing to correct errors encountered in long reads produced by SMRT sequencing (24). This hybrid approach has two steps: (i) SMRT read correction, where insertion/deletion errors present in SMRT read outputs are removed to produce corrected reads with sufficient accuracy and quality scores, and (ii) de novo assembly of the corrected SMRT reads, either independently or in conjugation with Illumina reads.
SMRT reads were corrected using the PacBioToCA package (http://sourceforge.net/apps/mediawiki/wgs-assembler/index.php?title=PacBioToCA) in wgs 7.0 (25, 26). The high-fidelity read set used for the correction was produced by using a combination of circular consensus SMRT reads, Illumina paired-end reads with sufficient overlap to merge into a single extended accurate read using fast length adjustment of short read (FLASH) (27), and Illumina paired reads without sufficient overlap for extension. Error correction resulted in a total of 570.1 Mbp, in 394,300 long corrected SMRT (C-SMRT) reads with an average phred quality score of 58.5 (28). These C-SMRT reads, which had an N50 of 1,686 bp and ranged between 500 bp and 10,932 bp in length, were subsequently used for de novo assembly. All sequence data included in the final assembly had an average quality score of 59.7. The final assembly was a marked improvement compared to the Illumina-only assembly, as evident from the improved N50/N90 values, the increase in the number of genes with PASA transcript alignment (see below), and the increase in the average length of gene models (see Table S2 in the supplemental material). More importantly, the long C-SMRT reads allowed for the identification of a large number of introns previously undetected using Illumina assembly; the extremely low GC content (8.1%) and the large number of microsatellites within these introns probably hindered their detection and assembly from short Illumina paired-end reads.
We used the core eukaryotic genes (CEGMA) to test the completion of the final assembly (29). Due to the unique nature of C1A, e.g., absence of gluconeogenesis, anaerobic fermentative mode of metabolism, and 22/454 genes being not expected to be present, we identified 408 out of 432 genes within the final assembly, suggesting a sequence completion of ≈94.4%.
RNA sequencing and gene calling. (i) RNA sequencing, assembly, and quantitative analysis.
RNA for RNA-seq analysis (30) was isolated from a log-phase strain C1A subculture grown and propagated on rumen fluid-free basal medium with cellobiose or cellulose using the Masterpure yeast RNA purification kit (Epicentre Corp., Madison, WI, USA). RNA sequencing libraries were generated using the Illumina TruSeq RNA sample protocol. Illumina sequencing was conducted using the services of commercial providers (Ambrys Genetics, Aliso Viejo, CA, USA, using a HiSeq 2000 sequencing platform using 100-bp paired-end chemistry for cellobiose treatment, and Centrillion Biosciences, Palo Alto, CA, using the Illumina Miseq platform for cellulose treatment). Transcripts generated on cellobiose were used in gene calling efforts as described below. The number of reads and Gbp produced per sequencing run is provided in Table S1 in the supplemental material. All quality-filtered reads were assembled into transcript candidates using the de novo transcriptome assembly program Trinity (31). The Trinity de novo assembly was executed using the standard non-strand-specific library settings with the addition of the “–jaccard_clip” option, which minimizes the formation of fusion transcripts, which are often present in fungal genomes, by checking for logical spatial orientation of paired reads on the assembled transcript. Transcripts with a base pair length greater than 300 bp were considered valid candidates for downstream analysis. Expression levels for the transcriptome assembly were calculated by mapping all pair-ended RNA-seq reads with bowtie (31) using the Trinity de novo assembly as the reference index. Quantitative expression estimates of alignment values were calculated in transcripts per million values with the RNA-Seq by Expectation-Maximization (RSEM) package (32). The cellobiose C1A transcriptome was utilized for gene calling through transcript alignment. However, while genomic data were used for genomic analysis in this study, in rare cases within pathway analysis (see Results below), transcriptome data were consulted in case a single gene or a few genes within a specific pathway were inexplicably missing from the genome.
(ii) Gene calling.
Contigs larger than 1,000 bp produced by the assembly were used as input contigs for gene model generation and downstream analysis. Gene calling was conducted using a combination of ab initio gene model prediction using GlimmerHMM (33) and Augustus (34) and transcript alignments from cellobiose treatment using PASA (Program to Assemble Spliced Assemblies) (35). Training parameters for the ab initio programs were generated using the de novo-assembled transcripts aligned with the genome assembly using GMAP (36). Additional gene hint parameters available for the Augustus program using the unassembled RNA-seq read data were generated using the software's recommended protocol (http://bioinf.uni-greifswald.de/augustus/binaries/readme.rnaseq.html).
Ab initio gene-calling algorithms produced 60,595 gene models, which were combined with 14,009 PASA high-quality transcript assemblies and 38,647 Trinity transcripts for genome GMAP alignments. The final single consensus gene model for each theoretical locus was produced by information-based source-weighted integration using EvidenceModeler (EVM) (35). A final total of 16,347 consolidated consensus gene models was generated by EVM (see Table S3 in the supplemental material).
To examine the impact of assembly fragmentation on the overall number of genes identified in the C1A genome, we extracted 300 bp from both ends of every contig in the C1A final genome assembly. To identify sequence redundancy, we queried these ends against NT and NR GenBank databases using BLASTN and BLASTX, respectively. The results show a unique first hit for each contig end, suggesting that the large number of contigs in the final assembly did not result in any false duplication of predicted genes. A similar result was obtained when performing the same analysis against the COG database.
Annotation. (i) Annotation strategy overview.
Annotation of gene models and gene transcripts was achieved using a combination of command line bioinformatics programs, manual curation, and automated online annotation suites. Closest-relative homologs were assigned to each consensus gene and de novo transcript model using the BLAST+ (37) module BLASTP for the gene models and BLASTX for putative transcripts against the NR database. Identified homologs with an E value of e−4 or less were considered sufficient for evaluation of functional activity assignment. PFAM domains were identified using the hmmscan module of the HMMER suite (38) against the PFAM 26.0 database of conserved protein families (39). All domains identified by hmmscan having a full-sequence E value of e−4 or less were assigned to the gene models for functional annotation. The integrated microbial genomes system (IMG) was used for automated gene calling and annotation of both the genome gene models and the transcripts. The resulting BLAST and IMG results were manually curated and used for analysis of various cellular processes within the genome, as well as for confirmation of the presence/absence of key metabolic genes using reciprocal BLASTP against the genome. All gene calling and annotation computational work were conducted using the Oklahoma State University high-performance computing clusters Pistol Pete and Spur.
(ii) CAZyme identification and analysis.
Identification of carbohydrate active enzyme (CAZyme) genes in the C1A genome, as well as in multiple genomes that were used for comparative analysis, was achieved via PFAM domain identification and analysis as described previously (40). The CAZyme database classification system (41) was used to classify glycoside hydrolase (GH), carboxyl esterase (CE), pectate lyase (PL), and carbohydrate-binding modules (CBMs). All models harboring a dockerin domain with an e−4 significance value or less were considered eligible for further analysis and classification. Potential secreted peptides and transmembrane proteins candidates were identified using SingalP 4.0 (42) and TMHMM (43).
Additionally, more-stringent controls were conducted to guard against any possible in silico gene number inflation within the CAZyme data set due to the fragmented assembly. To guard against in silico inflation of gene numbers due to improper assembly, we identified genes with nearly complete (>97%) amino acid identity. Each cluster of genes with nearly identical amino acid sequences was aligned, and genes with less than 80% alignment with the parent model were identified as possible artifacts and removed from the assembly. To guard against possible in silico inflation of gene numbers due to gene fragmentation between two contigs, we manually examined each CAZyme gene model to identify genes with an incomplete PFAM CAZyme domain. Genes with incomplete PFAM domains were removed from the assembly. Collectively, these additional quality control approaches resulted in the removal of 76 GH, 5 CE, and 5 PL genes.
(iii) Repeat identification.
Repetitive DNA sequences (DNA repeats) are defined as sequences present in more than a defined number of copies and that have no apparent biological function (44, 45). DNA repeats can be classified into simple sequence repeats (SSRs) and complex repeats. SSRs (also known as tandem repeats) are classified, depending on the length of the repeated unit, into microsatellites (basic unit length ranging from 1 to 6 bp) (44, 45) and minisatellites (basic unit length ranging from 15 to >150 bp repeated 2 to 100 times) (45). Longer tandem repeats constitute satellites (centromeres) and telomeric repeats. Complex repeats, on the other hand, result from transposable elements (TEs). TEs are further classified into class I retrotransposons (including long terminal repeats [LTRs] and non-LTRs) and class II DNA transposons.
Microsatellite (SSRs) in the C1A genome were identified using PHOBOS (C. Mayer, Ruhr-Universität Bochum, Bochum, Germany) with the minPerfection 100 flag to detect only perfect repeats. SSRs identified have the following minimum number of repeats: mononucleotide with at least 10 repeats; dinucleotides with at least 6 repeats; and tri-, tetra-, penta-, and hexanucleotides with at least 5 repeats.
Complex repeats were identified in the C1A genome as previously described (46) using a combination of RepeatScout (47), RepeatMasker (A. F. A. Smit, R. Hubley, and P. Green, 2003; http://www.repeatmasker.org), LTR_FINDER (48), and BLASTx against RepBase (http://www.girinst.org/repbase/index.html). Briefly, a consensus repeat library was created using the default parameters of RepeatScout. This library was filtered by removing short sequences (<100 bp) and those repeats with significant hits to Uniprot proteins (except repeats with significant hits to transposable elements). The filtered consensus library was then compared to the RepBase database using BLASTx for manual annotation and classification. LTRs were identified in the genome using LTR finder. Similar to other TEs, candidate LTRs were compared to the RepBase for classification. Finally, all candidate repeats classified by RepBase were used to mask the genome in RepeatMasker to identify the number of occurrences and the percent genome coverage of each TE class.
Gene duplication was identified by running local BLASTP using C1A proteins as both the subject and the query. Only the first and second hits were examined. The second hit is a protein with similarity to the query protein present anywhere in the genome. Percent similarity cutoffs of 40% or more were used.
(iv) Noncoding RNA identification.
rRNAs were identified using local BLASTN search with sequences corresponding to published 5.8S (5.8S fragment in GenBank accession number AJ864475.1), 18S (GenBank accession number AY546684.1; Spizellomyces punctatus), 28S (28S fragment in GenBank accession number AJ864475.1), ITS1 (GenBank accession number AF170191.1), and ITS2 (GenBank accession number JN943062.1) as the database. tRNAs were identified using tRNAscan-1.4 (49).
(v) Identification of proteases, protease inhibitors, and transporters.
All proteins were compared to the MEROPS database using BLASTP to identify potential proteases and protease inhibitors. Membrane transporters in the C1A genome were identified by BLASTP comparison against the transporter classification database (TCDB) sequences available at http://www.tcdb.org/seqfile/tcdb using the GBLAST2 program (http://www.tcdb.org/labsoftware.php).
(vi) Identification of hydrogenosomal proteins.
We bioinformatically predicted proteins potentially imported to the hydrogenosomal matrix in strain C1A using a combination of motif search and Mitoprot v1.0 (50). First, C1A proteins were examined for the presence of an N-terminal mitochondrial targeting sequence corresponding to a previously predicted motif similar to ML(S|T|A|C|G|R){0,1}X{0,19}RXF(I|L|F|S|A|G|Q), ML(S|T|A|C|G|R){0,1}X{0,19}R(F|N|E|S|G) (I|L|F|S|A|G|Q), MTLX{0,19}RXF(I|L|F|S|A|G|Q), MTLX{0,19}R(F|N|E|S|G) (I|L|F|S|A|G|Q), MSLX{0,19}RXF(I|L|F|S|A|G|Q), or MSLX{0,19}R(F|N|E|S|G) (I|L|F|S|A|G|Q), where X is any amino acid except tryptophan. Numbers between braces refer to the previous residue repeat number, and parentheses indicate that any of the residues enclosed is possible at that position (7). Mitochondrial import probabilities of proteins harboring this N-terminal motif were then predicted using Mitoprot v1.0 (50), where an arbitrary probability of 0.6 was used as the cutoff. Using these criteria, we identified 21 potential intrahydrogenosomal proteins. Further, Mitoprot was also used to predict the mitochondrial import probabilities of proteins with similarity to known mitochondrial matrix proteins that did not have the above mitochondrial-targeting motif. An additional 25 potential hydrogenosomal proteins were identified using these criteria.
Comparative analysis of the C1A genome to basal fungi, Dikarya, and Opisthokonta genomes.
We used local BLASTP comparison of C1A proteins against all Mycota proteins, as well as against Dikarya proteins. A Mycota BLASTP database was created by downloading proteins of all sequenced fungal genomes available from GenBank and IMG (total of 116 fungal genomes). Of those, 4 belonged to early-branching lineages (Allomyces macrogynus, Batrachochytrium dendrobatidis, Spizellomyces punctatus, and Rhizopus oryzae). The remaining fungal genomes constitute the Dikarya BLASTP database. The number and identity of BLASTP first hits of C1A proteins against both databases at different E value cutoffs (e−5, e−10, e−15, e−20, e−25, e−30, and e−35) were then used to specify C1A proteins that are general fungal proteins (present in both early-branching and Dikarya fungi), early-branching fungus-specific proteins (present only in early-branching fungi but not in Dikarya fungi), and C1A-specific proteins (specific to C1A and absent from other fungal genomes). The last (C1A-specific proteins) were compared to the NR database excluding Mycota using the flag –negative_gilist. Proteins with no hits in the NR database were considered C1A hypothetical proteins. Functional annotation of various C1A-specific and early-branching fungus-specific proteins identified was conducted using the PANTHER classification outline (51).
Lignocellulolytic capabilities of strain C1A. (i) Plant materials and pretreatment.
Samples of mature Kanlow switchgrass (Panicum virgatum var. Kanlow), mature Sorghum bicolor, and mature energy cane (Saccharum officianarum var. Ho02) were obtained from Oklahoma State University experimental plots in Stillwater, OK. Dried alfalfa was obtained from a local farm and ranch supplier. Samples of Bermuda grass (Cynodon dactylon) were obtained from residential lawn clippings in Guthrie, OK. Samples of corn stover from Zea mays were obtained from the Industrial Agricultural Products Center at the University of Nebraska in Lincoln. Untreated wood samples, including cedar (Juniperus sp.), oak (Quercus sp.), and pine (Pinus sp.), were obtained from a local lumberyard in Stillwater, OK. Cottonwood (Populus deltoides) and willow (Salix babylonica) wood samples were harvested from live trees growing in the Stillwater area. All samples were dried at 45°C overnight, milled, and sieved to a final particle size of 2 mm as previously described (52).
Sodium hydroxide (NaOH) treatments were conducted by heating 4 g of dried plant material in 40 ml of a 1% NaOH solution inside a sealed serum bottle at 50°C for 12 h (53). Acid treatment was conducted by heating 4 g of dried plant material in 40 ml of 0.5% H2SO4 inside a sealed serum bottle for 1 h (54). Hydrothermolysis-treated switchgrass was prepared by mixing 60 g of switchgrass with distilled water to achieve a 10% dry matter mixture (52). This mixture was placed inside a 1-liter benchtop pressure reactor (Parr Series 4520; Parr Instrument Company, Moline, IL, USA) that was heated to 200°C and agitated at 500 rpm (52). The switchgrass-water mixture was held at 200°C for 10 min and then cooled in an ice bath (52). All of the treated switchgrass samples were recovered from pretreatment incubations by filtration. The sodium hydroxide- and acid-treated switchgrass was washed with deionized water as previously described (53, 54). All of the pretreated switchgrass samples were dried at 45°C for approximately 48 h before they were used in the experiments described below.
(ii) Growth of strain C1A on plant materials.
Experiments to evaluate the growth of strain C1A on different treated and pretreated plant materials were conducted under strict anaerobic conditions in 160-ml serum bottles. All experiments were conducted in triplicate, and unless otherwise specified, 0.5 g of plant material was used as the substrate. Experiments were conducted in a previously described rumen fluid-free basal medium (17). The medium was prepared under strict anaerobic conditions using 100% CO2 and the techniques of Bryant (18), as modified by Balch and Wolfe (19). Once the basal medium was prepared, it was autoclaved for 20 min at 121°C and 15 lb/in2 of pressure and then cooled. Each serum bottle was then amended with the appropriate type of plant biomass inside an anaerobic chamber (Coy Laboratory Products, Grass Lake, MI). After the serum bottles were amended with plant materials, they were removed from the glove bag and the headspace was repressurized with 15 lb/in2 of 100% CO2 (19). Five milliliters of an actively growing culture of strain C1A (approximately 2.6 mg of fungal biomass) was used as an inoculum and added to 45 ml medium in 160-ml serum bottles. In all experiments, serum bottles were incubated at 39°C in a nonshaking incubator. Substrate-unamended controls were included in all experiments to account for any product carryover from the inoculum. Triplicate bottles were sacrificed at different time intervals to quantify substrate loss and product formation.
(iii) Analytical methods.
Fatty acids and ethanol in supernatant fractions were quantified using a high-pressure liquid chromatograph (HPLC) with a refractive index detector (1100 series; Agilent, Santa Clara, CA, USA) and an Aminex HPX-87H column (Bio-Rad, Sunnyvale, CA, USA), which was heated to 60°C. The mobile phase was 0.01 N H2SO4, with a flow rate of 0.6 ml per minute. Sugars in supernatant fractions were also quantified using an HPLC with a refractive index detector (1100 series; Agilent, Santa Clara, CA, USA). The HPLC was equipped with an Aminex HPX-87P column (Bio-Rad, Sunnyvale, CA, USA), which was heated to 85°C. Distilled water was used as the mobile phase at a flow rate of 0.6 ml per minute.
The amount of plant material consumed in serum bottles was calculated by subtracting the time final from the time zero dry weights of each plant material. Since the time final pellets contained a mixture of plant and fungal biomass, the amount of fungal biomass at time final was indirectly quantified using formate concentrations as previously described (12). The amounts of cellulose, xylan, hemicellulose, and lignin in the different plant substrates were determined using the standard NREL procedures (55). The procedure included the addition of 3 ml of 72% sulfuric acid to each sample and incubation at 30 ± 3°C for 1 h, with stirring every 5 to 10 min. The samples were then diluted with 84 ml of deionized water, capped, and autoclaved for 1 h to 121°C. The cooled solution was filtered, and this filtrate was used to determine carbohydrate content and soluble lignin. The remaining solids were washed and dried to constant weight at 105°C to determine acid-insoluble residue (AIR) and then converted to ash at 575°C for 24 h (55). Analyses of resulting carbohydrates within the filtrate were done by HPLC with refractive index detection (RID) (Agilent 1100 series; Santa Clara, CA) on an Aminex HPX-87P column at 85°C with a mobile phase of deionized water pumped at 0.6 ml/min for 30 min (55). Twenty microliters of each sample was analyzed for cellobiose, glucose, xylose, galactose, arabinose, and mannose. Contributions of structural constituents to the total biomass composition were determined using the NREL summative mass closure procedure (56). The acid-soluble lignin (ASL) content was determined using a UV spectrophotometer set at a wavelength of 205 nm, as has been previously used to determine ASL in switchgrass (57). As recommended in the NREL procedure, ASL in corn stover was measured at 320 nm, whereas a 240-nm wavelength was used for the remaining biomass types (55).
Nucleotide sequence accession numbers.
The final genome assembly is available in the IMG genome database with accession number 2518645524 and in the GenBank database with accession number PRJNA200719. The final transcriptome is available in the IMG database with accession number 2510461071.
RESULTS
Isolation and general genomic features.
Strain C1A was isolated from the feces of an Angus steer on a cellobiose-switchgrass medium using previously described protocols (16). The isolate displayed polycentric growth and effectively colonized switchgrass. Phylogenetic analysis using the nuclear ribosomal internal transcribed spacer II (ITS-II) region supported the placement of strain C1A as a member of the genus Orpinomyces, and phylogenetic analysis using a concatenated set of 42 housekeeping genes supported the basal, early-diverging position of the Neocallimastigomycota (see Fig. S1 in the supplemental material).
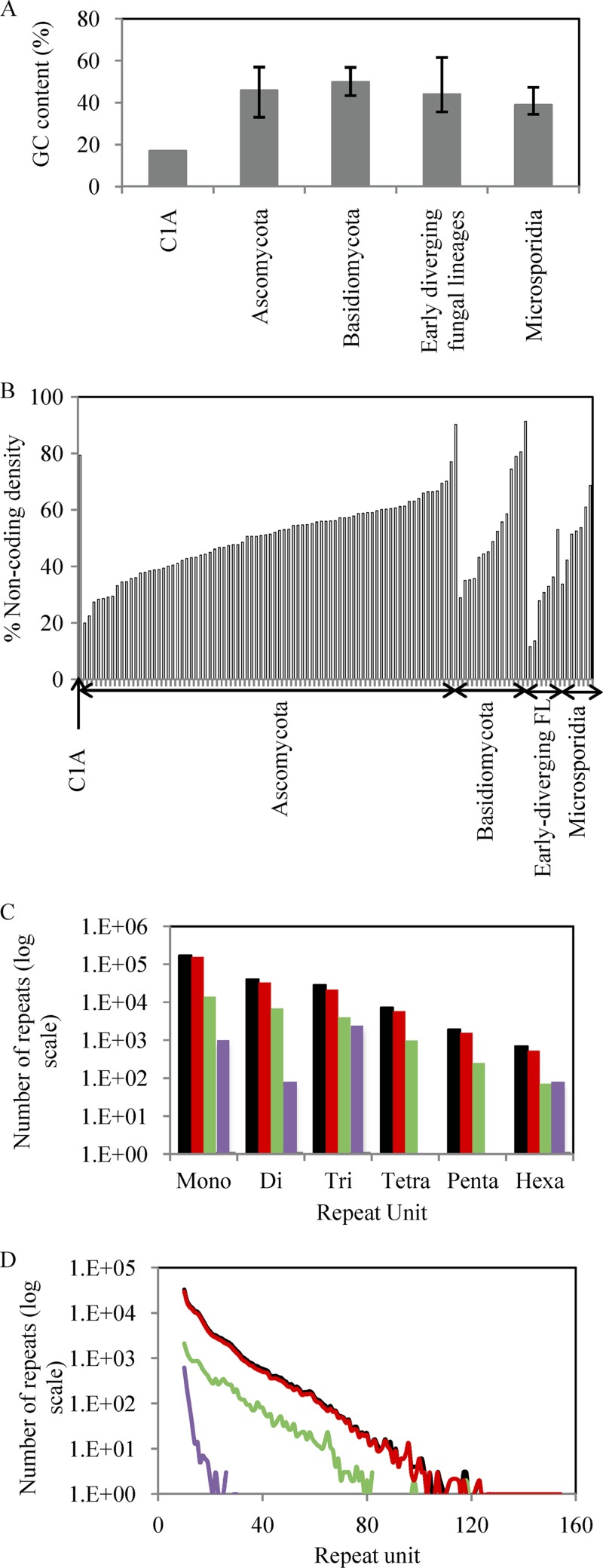
We sequenced the C1A genome using a combination of paired-end short-read Illumina technology (≈290× coverage) and single-molecule real-time (SMRT) Pacific Biosciences technology (≈10× coverage). The transcriptomes of strain C1A grown on cellobiose and cellulose were also sequenced using Illumina technology (see Tables S1 and S2 in the supplemental material). The C1A genome displayed several interesting features (Fig. 1; see also Fig. S2 and S3 and Tables S4 to S8). It had the lowest GC content (17.0%) compared to available genomes of all free-living microorganisms sequenced to date (Fig. 1A). This value is lower than those observed in the notoriously AT-rich Dictyostelium spp. and Plasmodium sp. within the microeukaryotes and is surpassed only by a few proteobacterial obligate endosymbionts, e.g., “Candidatus Zinderia insecticola” (13.5%) and “Candidatus Carsonella ruddii” (16.6%). The GC content was higher in protein-coding genes (26.8%) than in noncoding regions (14.8% in intergenic regions and 8.1% in introns) but still resulted in a marked codon usage skew (see Table S6). The C1A genome was also characterized by a relatively large proportion of noncoding intergenic regions (73.1%) (Fig. 1B). Further, noncoding regions displayed massive proliferation of simple sequence repeats (SSRs) in the C1A genome. The 249,194 SSRs constituted 4.9% of the entire genome, as well as 5.8% and 6.0% of the intergenic region and the introns, respectively (Fig. 1C). These values vastly surpass the number of SSRs that were observed in previously analyzed fungal genomes by at least 1 order of magnitude (46, 58). Homopolymeric A or T monorepeats represented the majority of observed repeats (68.6% and 60.6% of total SSR numbers and length, respectively), with 3,589 identified cases of ≥50-bp stretches of A or T within the assembly (Fig. 1D).
Fig 1.
Unique features in the Orpinomyces sp. strain C1A genome. (A) The C1A genome has the lowest G+C content in all fungal genomes described thus far. Averages and ranges were computed from the publicly available genomes of Ascomycota (n = 90), Basidiomycota (n = 16), basal fungal lineages (n = 6), and Microsporidia (n = 7). (B) The C1A genome has large intergenic noncoding regions compared to publicly available fungal genomes. A list of 110 genomes for comparison is available in Table S5 in the supplemental material. (C) The C1A genome has the highest recorded abundances of simple sequence repeats within the Mycota, with the majority of repeats in intergenic regions and introns. (D) The homopolymeric (A/T) mononucleotide repeats in the C1A genome not only were abundant but also reached lengths of up to 151 bp in intergenic regions. Color codes: black, genome; red, intergenic region; green, introns; purple, cDNA.
Comparative gene content with basal and Dikarya fungi.
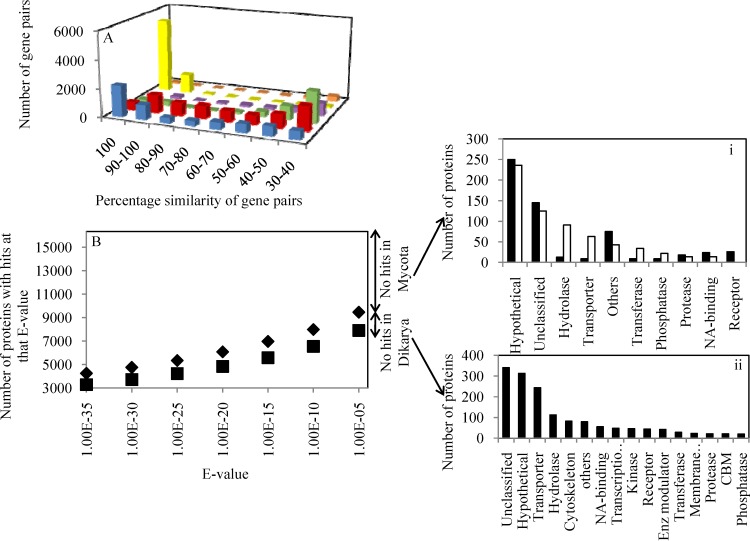
Gene calling resulted in the identification of 16,347 protein-coding genes, a number surpassed by only a few fungal genomes (see Text S1 and Table S5 in the supplemental material). This large number could partly be attributed to gene duplication (Fig. 2A), since 3,252 gene pairs share >90% sequence similarity.
Fig 2.
(A) Gene duplication in C1A genome compared to other fungal genomes. Color codes: blue, C1A; red, Laccaria bicolor; green, Magnaporthe grisea; purple, Saccharomyces cerevisiae; yellow, Candida albicans; orange, Encephalitozoon cuniculi. (B) Identification of C1A genes with at least one ortholog within Dikarya (n = 116) (■) or Mycota (Dikarya plus basal fungi; n = 4) (◆) genomes at different E value cutoffs. (i) Panther classification (51) and putative phylogenetic affiliation based on BLAST first hit of C1A genes not encountered in Mycota (black bars, nonfungal eukaryotes; open bars, prokaryotes) (i) and genes encountered only in basal fungi (ii).
In addition, comparative genomic analysis indicated that only 48.4% of C1A genes have at least one ortholog in all examined Dikarya genomes (n = 116), that 9.5% of C1A genes have at least one ortholog within examined early-branching fungal genomes (n = 4) but not in Dikarya genomes, and that 42.2% (6,886) of C1A genes are unique and have not been previously encountered within the Mycota (Fig. 2B; see also Table S9 in the supplemental material), These unique C1A genes were either genes for hypothetical proteins (n = 5,666); genes with nonfungal, eukaryotic orthologs (n = 578); or genes with bacterial orthologs (n = 642). Eukaryotic, nonfungal C1A genes were mostly encoding cellular processes, e.g., receptors and nucleic acid-binding proteins, highlighting the distinct early-branching fungal position of the Neocallimastigomycota, while C1A genes with prokaryotic orthologs were mainly involved in metabolic processes, e.g., hydrolases, transporters, transferases, and phosphatases, highlighting the potential role of horizontal gene transfer in shaping C1A metabolic capabilities.
Genomic analysis and comparative genomics reveal multiple differences between Neocallimastigomycota and Dikarya.
Detailed analysis of metabolic capabilities, cellular processes, and structural features in the C1A genome is presented as supplemental material (see Text S1 and Fig. S4 in the supplemental material). Briefly, analysis of genes involved in information processing (replication, transcription, and translation), as well as cytoskeletal structure and intracellular trafficking mechanisms, revealed all salient features associated with such processes in eukaryotic cells.
More importantly, comparative genomic analysis identified multiple cellular processes in which either only the C1A genome or all early-branching fungal genomes possess features that appear to be absent from Dikarya genomes but are mostly associated with nonfungal Opisthokonta (choanoflagellates such as Monosiga brevicollis and Capsaspora owczarzaki, an independent unicellular Opisthokonta lineage, and Metazoa) and higher non-Opisthokonta eukaryotes (Table 1; see Text S1 and Tables S10 and S11 in the supplemental material). Five different examples are highlighted. (i) One gene in the C1A genome encodes a member of the metalloprotease site-2-protease (S2P) family, and seven different genes encode various components of the γ-secretase complex, including aspartyl protease presenelin. Both of these types of intramembrane proteases are represented in Metazoa (mostly Chordata, Nematoda, and Arthropoda), with a few representatives in plants, and have no representation in Dikarya (Table 1). (ii) The genome contains two fucosyltransferase genes that mediate fucosylation, a posttranslational modification process that is typically observed in Chordata, Arthropoda, and Viridiplantae but not in the Dikarya (Table 1). (iii) The C1A genome possesses a near-complete focal adhesion (FA) machinery (Table 1). FAs are large multiprotein intracellular assemblies that mediate cell anchorage and mechanical adhesion to the extracellular matrix. They also act as a signaling milieu where signaling proteins are concentrated at sites of integrin binding and connect the cell's cytoskeleton to the extracellular matrix. FA appears to be absent from filamentous fungi and more common in other eukaryotes (Amoebozoa, Metazoa [sponges, placozoans, and cnidarians], and Holozoa) (59). (iv) The C1A genome possesses a complete axoneme and intraflagellar trafficking machinery proteins. The axoneme acts as a scaffold for other protein complexes, including motor proteins (e.g., kinesin and dynein) essential for intraflagellar transport of proteins. Ciliated and flagellated eukaryotic cells are known to possess an axoneme, as do Neocallimastigomycota and other early-branching fungi that produce flagellated zoospores. This feature is absent in other Dikarya fungi that produce nonflagellated spores. (v) Finally, the C1A genome encodes various extracellular protease inhibitors, some of which (serpins) have not previously been encountered in the Dikarya (e.g., serpins of family I4 are present mostly within eukaryotic metazoan phyla [Arthropoda, Chordata, and Nematoda] and have also been identified in Bacteria and Archaea but have not previously been encountered in fungi). Several identified serpins have dockerin domains, confirming their cellulosomal destination and their potential role in combating plant proteases, as previously suggested (60).
Table 1.
Salient differences identified between strain C1A genome, basal fungal genomes, Dikarya fungal genomes, and unicellular Opisthokonta genomes
| Category | Gene identifier | Product name | Present in: |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Basal fungi |
Dikarya fungi | Opisthokonta |
|||||||
| Allomyces macrogynus ATCC 38327 | Batrachochytrium dendrobatidis JEL423 | Spizellomyces punctatus DAOM BR117 | Rhizopus oryzae RA 99-880 | Monosiga brevicollis | Capsaspora owczarzaki | ||||
| Intramembrane proteases | |||||||||
| Gamma secretase complex | 2510864531 | Aph-1 protein | Yes | Yes | Yes | Yes | No | Yes | Yes |
| Orpinomyces_14795 | Nicastrin | No | Yes | Yes | Yes | No | Yes | Yes | |
| Orpinomyces_14978 | Nicastrin | No | Yes | Yes | Yes | No | Yes | Yes | |
| Orpinomyces_3406 | Nicastrin | No | No | No | No | No | No | No | |
| 2510858953 | Presenilin enhancer 2 | No | No | No | No | No | No | Yes | |
| Orpinomyces_8327 | Presenilin | Yes | Yes | Yes | Yes | No | Yes | Yes | |
| Orpinomyces_13802 | Presenilin | Yes | Yes | Yes | Yes | No | Yes | Yes | |
| Site 2 peptidase | Orpinomyces_8676 | Peptidase family M50A | No | No | Yes | No | No | No | No |
| Posttranslational protein modification | Orpinomyces_8992 | GT family 10 (fucosyltransferase) | No | No | No | Yes | Noa | No | No |
| Orpinomyces_15723 | GT family 10 (fucosyltransferase) | No | No | No | Yes | Noa | No | No | |
| Focal adhesion proteins | |||||||||
| Adaptor proteins | 2510867334 | Paxillin | No | No | No | Yes | No | Yes | Yes |
| Orpinomyces_16019 | Talin | No | No | No | No | No | Yes | Yes | |
| Orpinomyces_11547 | Band 4.1/talin | No | Yes | Yes | No | No | Yes | Yes | |
| Orpinomyces_13613 | Vinculin family | No | No | No | No | No | No | No | |
| Orpinomyces_13888 | Vinculin family | No | No | Yes | No | No | Yes | No | |
| Orpinomyces_8561 | Vinculin family | No | Yes | Yes | No | No | Yes | Yes | |
| IPP complex components | Orpinomyces_13334 | Integrin-linked kinase | No | No | Yes | No | No | No | Yes |
| Orpinomyces_16310 | Integrin-linked kinase | No | Yes | Yes | No | No | No | Yes | |
| 2510867610 | α-Parvin | No | No | Yes | No | No | No | Yes | |
| Orpinomyces_13214 | PINCH-1 | Yes | Yes | Yes | Yes | No | Yes | Yes | |
| Downstream-acting elements | Orpinomyces_3201 | ROCK | No | Yes | Yes | No | No | Yes | No |
| Axonemal proteins | Several (43) proteins were identified in the C1A genome and are thought to be involved in axoneme structure and function; details are shown in Table S10 in the supplemental material | Someb | Someb | Someb | Someb | No | Somec | Somec | |
| Protease inhibitors | |||||||||
| Serine proteases (serpins) family I4 | Orpinomyces_14234 | Protease inhibitor I4 | No | No | No | No | No | No | No |
| Orpinomyces_14684 | Protease inhibitor I4 | No | No | No | No | No | No | No | |
| Orpinomyces_6138 | Protease inhibitor I4 with dockerin domain | No | No | No | No | No | No | No | |
| Orpinomyces_2311 | Protease inhibitor I4 with dockerin domain | No | No | No | No | No | No | No | |
| Apoptosis inhibitors (BIR domain) cysteinyl protease inhibitors family I32 | Orpinomyces_4452 | Apoptosis inhibitor and related BIR domain proteins | No | No | Yes | Yes | Yesd | No | No |
| Cysteinyl protease inhibitor family I42 | 2510865883 | Chagasin | Yes | No | No | No | No | No | No |
| 2510871764 | Chagasin | No | No | No | No | No | No | No | |
| 2510864005 | Chagasin | No | No | No | No | No | No | No | |
In all Dikarya genomes currently available in the GenBank database, only one gene annotated as “hypothetical protein” in Puccinia graminis (EFP84060) with low sequence similarity to plant fucosyltransferases has been identified.
Homologs for some of the C1A axonemal proteins were found in the basal fungal genomes. None of the BBSome proteins were detected in any of the fungal genomes. Details are shown in Table S10 in the supplemental material.
Homologs for axonemal proteins were detected before in Opisthokonta representatives.
Homologs are found only in Coccidioides immitis RS, Coccidioides posadasii C735 delta SOWgp, Coprinopsis cinerea okayama7#130, Fusarium oxysporum lycopersici FGSC 4286, Gibberella zeae PH-1, Nosema ceranae BRL01, Phanerochaete chrysosporium RP-78, Saccharomyces cerevisiae AWRI1631, Saccharomyces cerevisiae YJM789, Saitoella complicata NRRL Y-17804, Scheffersomyces stipitis CBS 6054, and Ustilago maydis 521.
Hydrogenosomal structure and function.
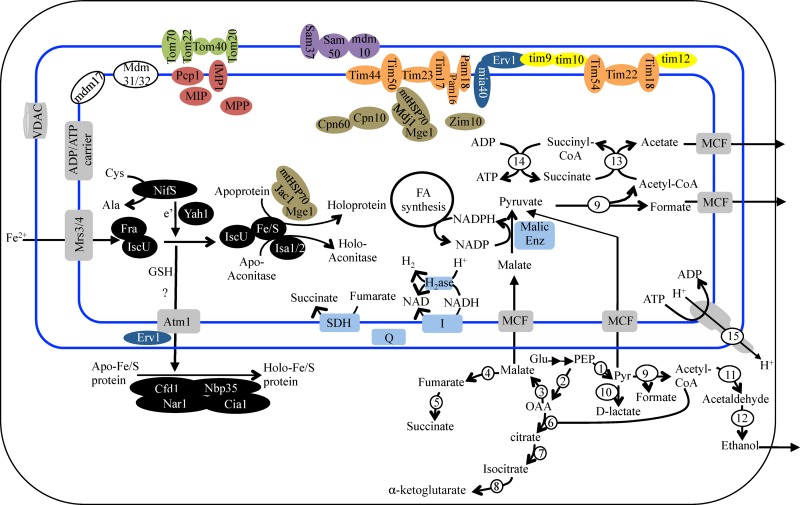
Anaerobic fungi lack mitochondria but possess a double-membrane hydrogenosome whose main function is ATP production via substrate-level phosphorylation and hydrogen production (6, 8, 9). The C1A genome encodes a near-complete hydrogenosomal protein import system with components of the TOM outer membrane transport system (4 out of 7 genes), the SAM sorting and assembly complex (4 out of 5 genes) for protein insertion in the outer membrane, the MIA intermembrane space import and assembly complex (2 out of 3 genes), small TIMs (2 out of 4 genes), the TIM22 complex for protein insertion in the inner membrane (6 out of 6 genes), and the inner membrane transport system and associated motor (TIM23 complex, 10 out of 11 genes) (Fig. 3). In comparison, the hydrogenosomal import machinery of Trichomonas vaginalis has been reduced to a few outer membrane proteins (Tom40, Sam50, Hmp35, and Hmp36), a few inner membrane proteins (Tim17/22/23, Tim44, PAM16, and PAM18), and one highly modified intermembrane small TIM (61). Further, examining the phylogenetic affiliation of mitochondrial import proteins in strain C1A clearly demonstrates their fungal origin, since their closest relatives are consistently those from fungal mitochondria.
Fig 3.
Reconstruction of C1A hydrogenosome from genomic data. The double blue lines depict the hydrogenosomal outer and inner membrane. Various functional protein groupings are color coded: outer mitochondrial membrane translocase complex components (TOM) are shown in green, outer membrane sorting and assembly complex components (SAM) are shown in purple, inner membrane complex components (TIM) are shown in orange, intermembrane space import and assembly proteins (MIA) are shown in blue, intermembrane space small TIMs are shown in yellow, mitochondrial peptidases (inner membrane peptidase [IMP], mitochondrial processing peptidase [MPP], mitochondrial intermediate peptidase [MIP], and the mitochondrial signal peptidase PCP1) are shown in red, mitochondrial distribution and morphology (MDM) proteins are shown in white, chaperones and cochaperones are shown in brown, Fe-S assembly proteins are shown in black, and membrane transporters are shown in gray (VDAC, voltage-dependent anion channel; MCF, mitochondrial carrier family). Enzymes involved in pyruvate metabolism, substrate-level phosphorylation, and redox carrier regeneration are numbered: 1, pyruvate kinase; 2, phosphoenolpyruvate (PEP) carboxykinase; 3, malate dehydrogenase; 4, fumarase; 5, fumarate reductase; 6, citrate synthase; 7, aconitase; 8, isocitrate dehydrogenase; 9, pyruvate formate lyase; 10, d-lactate dehydrogenase; 11, acetaldehyde dehydrogenase; 12, alcohol dehydrogenase; 13, acetyl-CoA hydrolase (acetyl-CoA:succinyl transferase); 14, succinyl-CoA synthase; 15, ATP synthase. SDH, succinate dehydrogenase; H2ase, hydrogenase; I, complex I NADH dehydrogenase; Q, quinone.
Using two different bioinformatic criteria, we identified 46 intrahydrogenosomal proteins in the C1A genome. Candidate proteins for import into the hydrogenosomal matrix included several hypothetical proteins, Fe-S cluster assembly and maturation proteins, peptidases, and intraluminal chaperones and cochaperones, as well as pyruvate metabolism and energy production enzymes (see Tables S12 to S14 in the supplemental material).
Hydrogenosomes are the sites of multiple metabolic processes for pyruvate metabolism, ATP production via substrate-level phosphorylation, and regeneration of reduced electron carriers, e.g., NADPH and NADH. The C1A genome carries the genes required for mixed-acid fermentation, the predominant pathway for pyruvate metabolism in anaerobic fungi, as previously suggested (62–64) (Fig. 3; see also Table S13 in the supplemental material). Genomic reconstruction suggests that pyruvate produced from sugar catabolism in the cytosol could be either metabolized cytosolically or imported and metabolized in the hydrogenosome. In the cytosol, pyruvate could be either converted to acetyl coenzyme A (acetyl-CoA) and formate via cytosolic pyruvate formate lyase (PFL) (a cytosolic PFL-activating enzyme is also encoded by the genome), converted to d-lactate via cytosolic d-lactate dehydrogenase, or used to produce tricarboxylic acid (TCA) intermediates required for anaplerotic reactions via an incomplete cytosolic TCA cycle. In the cytosol, acetyl-CoA produced could be converted to ethanol via aldehyde dehydrogenase/alcohol dehydrogenase. In the hydrogenosome, pyruvate could be imported from the cytosol or could be produced from malate via the action of the hydrogenosomal malic enzyme (with the production of CO2). Hydrogenosomal pyruvate could be then metabolized to acetyl-CoA and formate by a hydrogenosomal PFL. Acetyl-CoA produced in the hydrogenosome could subsequently be converted to acetate via the combined action of hydrogenosomal acetate:succinate CoA transferase/succinyl-CoA synthase to produce ATP via substrate-level phosphorylation. The genome encodes a hydrogenosomal acetyl-CoA hydrolase. A similar enzyme in T. vaginalis was shown to possess an acetate:succinate CoA transferase activity. The Orpinomyces acetyl-CoA hydrolase homolog is most likely performing a similar transferase activity that, coupled to the succinyl-CoA synthase activity, could convert acetyl-CoA to acetate.
In addition, the hydrogenosomal components contain elements for NADH recycling coupled to H2 production (Fe-only hydrogenase large subunit and NADH dehydrogenase [complex I] subunits E and F). The 2 subunits of NADH dehydrogenase most probably function to reoxidize NADH produced in the lumen (e.g., during fatty acid degradation) and transfer electrons to Fe-only hydrogenase. Since PFL mediates pyruvate metabolism without the production of reduced equivalents, H2 production via the hydrogenase enzyme is thought to be minor and to be required only to cope with the NADH produced from other intraluminal reactions, e.g., 3-hydroxyacyl-CoA dehydrogenase of fatty acid metabolism (65). Elements of hydrogenosomal NADPH recycling are also present in the genome. NADPH produced from NADP-dependent reactions, e.g., malic enzyme, could possibly be used by the NADP-requiring fatty acid synthesis reactions, e.g., 3-oxoacyl-[acyl-carrier protein] reductase, or recycled by NADPH:quinone reductase to a quinone, where electrons could then transfer to succinate dehydrogenase to reduce fumarate to succinate. Finally, the genome also encodes subunits α, β, γ, and δ of FoF1-type ATP synthase that is thought to pump protons to the cytosol, keeping the luminal pH slightly alkaline. ATP synthase is likely functioning in conjunction with an ADP/ATP carrier.
Lignocellulolytic repertoire of strain C1A.
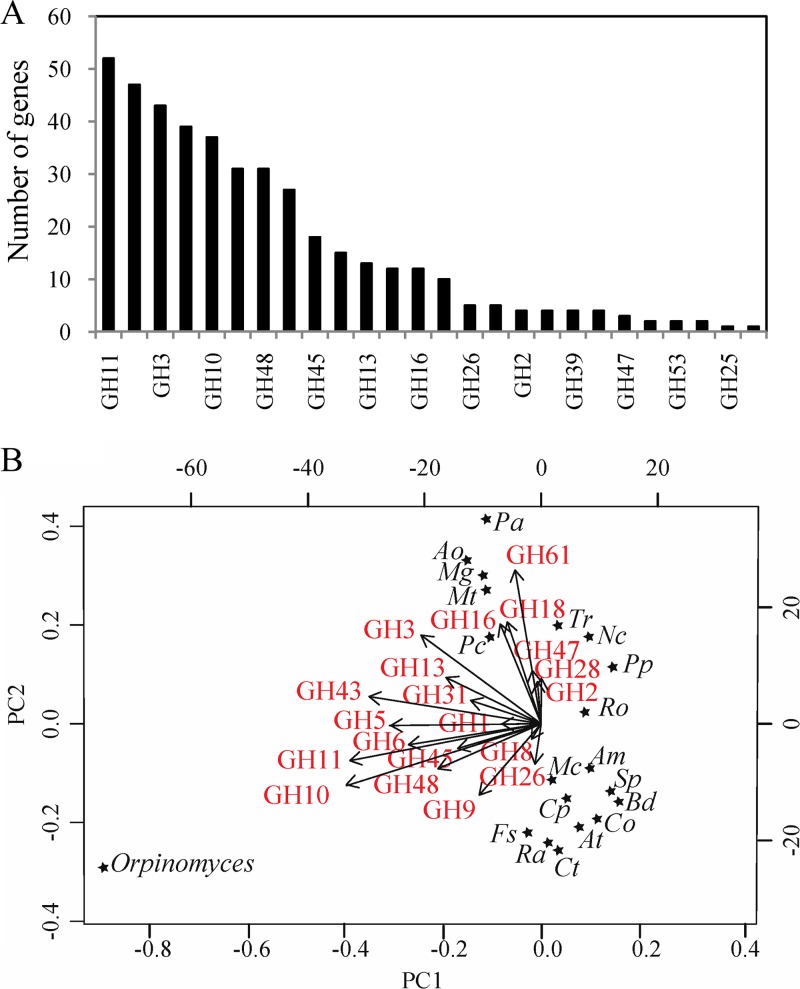
Prior research efforts have identified multiple genes involved in plant biomass degradation in several Neocallimastigomycota isolates (5, 14, 15, 66–88). To provide an overall view of the plant biomass degradation machinery of an anaerobic rumen fungus, we analyzed the lignocellulolytic machinery in the C1A genome. Such an analysis revealed an extremely rich repertoire that consisted of 357 glycoside hydrolase (GH) genes, 24 polysaccharide lyases (PLs), and 92 carbohydrate esterases (CEs) (Fig. 4A; see also Tables S18 to S20 in the supplemental material). Principal-coordinate analysis demonstrated the unique position of the GH catalytic machinery, compared to multiple fungal and bacterial genomes of distinct habitats, ecological roles, phylogenetic affiliation, and oxygen preferences (Fig. 4B). For instance, compared to aerobic fungal biomass degraders of industrial and ecological relevance such as Trichoderma reesei, Postia placenta, Aspergillus oryzae, and Myceliophthora thermophila, the C1A genome shows an expansion of cellulolytic families GH6, GH9, GH45, and GH48 and hemicellulolytic families GH10, GH11, and GH43, as well as the reduction or absence of families GH7, GH16, GH18, GH28, and GH61.
Fig 4.
Glycoside hydrolase (GH) families in the C1A genome. (A) Number of C1A genes belonging to different GH families. (B) Principal-coordinate analysis biplot of the distribution of GH families in the C1A genome, compared to those in 19 other selected fungal and bacterial genomes. Genomes are represented by stars, and GH families are represented by arrows. The arrow directions follow the maximal abundance, and their lengths are proportional to the maximal rate of change between genomes. Am, Allomyces macrogynus; At, Anaerocellum thermophilum DSM 6725; Ao, Aspergillus oryzae; Bd, Batrachochytrium dendrobatidis; Co, Caldicellulosiruptor obsidiansis; Cp, Clostridium phytofermentans ISDg; Ct, Clostridium thermocellum ATCC 27405; Fs, Fibrobacter succinogenes subsp. succinogenes S85; Mg, Magnaporthe grisea; Mc, Mucor circinelloides; Mt, Myceliophthora thermophila; Nc, Neurospora crassa; Pa, Podospora anserina; Pp, Postia placenta; Ro, Rhizopus oryzae; Ra, Ruminococcus albus 7; Sp, Spizellomyces punctatus; Tr, Trichoderma reesei.
Detailed phylogenetic analysis (see Fig. S5 in the supplemental material) suggests that the GH machinery in strain C1A has evolved from an ancestor with relatively limited cellulolytic capability to a robust cellulolytic and hemicellulolytic organism through the acquisition of genes from multiple bacterial lineages, many of which are known to be prevalent in the bovine rumen. Overall, 247 (69.2%) of GH genes were most closely related to bacterial orthologs, and 141 (39.5%) of GH genes were most closely related to bacterial orthologs from lineages that are prevalent in the bovine rumen. Such lineages include the families Lachnospiraceae, Clostridiaceae, Eubacteriaceae, and Ruminococcaceae within the order Clostridiales, the family Streptococcaceae within the order Bacillales, and the family Prevotellaceae within the order Bacteroidales, as well as the phylum Fibrobacteres. Cellulose degradation machinery in strain C1A consists of GH5, GH8, GH9, and GH45 endoglucanases and GH6 and GH48 cellobiohydrolases. GH8 is an exclusively prokaryotic gene family (41), and phylogenetic analysis of GH5 and GH9 endoglucanases indicates their close affiliation with endoglucanases from multiple bacterial sources, including the ruminal genera Clostridium, Ruminococcus, and Eubacterium. On the other hand, strain C1A also possesses the distinctively eukaryotic fungus-affiliated GH45 endoglucanases that have rarely been observed in bacterial genomes. Similar to endoglucanases, a dual prokaryotic/eukaryotic origin of strain C1A cellobiohydrolases was observed. Strain C1A possesses multiple GH48 reducing-end cellobiohydrolases, hallmarks of cellulosomal cellobiohydrolases, as well as multiple GH6 non-reducing-end cellobiohydrolases that are common in fungal genomes but rarely observed in anaerobic cellulolytic bacteria.
Unlike cellulose metabolism, hemicellulose degradation machinery in strain C1A appears to be entirely of prokaryotic origin. The C1A genome contains all genes required for the degradation of xylans (glucuronoarabinoxylans and arabinoxylans), mannans (galactoglucomannans and glucomannans), and mixed β-(1,3-1,4) glucans. Strain C1A appears to be highly adapted to the degradation of xylans, the prevalent hemicelluloses in grasses (order Poales) (89). This is evident by the identification of 109 different xylanases, xylosidases, arabinofuranosidases, and α-glucuronosidases belonging to families GH10, GH11, GH39, GH43, and GH67, in addition to multiple glucuronoarabinoxylan- and arabinoxylan-debranching enzymes (acetylxylan esterases, ferulic acid esterases, and polysaccharide deacetylases). Phylogenetic analysis of GH10 xylanases suggests their close affiliation with multiple bacterial lineages, including the ruminal genera Butyrivibrio, Clostridium, and Eubacterium. Phylogenetic analysis of GH11 xylanases suggests that they have been solely acquired from Fibrobacter species, important constituents of rumen microbiota. A similar bacterial origin was also observed for GH39 and GH43 xylosidase/arabinofuranosidases as well as GH67 α-glucuronosidases, with potential bacterial donors being members of the genera Clostridium, Ruminococcus, Butyrivibrio, Cellulosilyticum, Eubacterium, and Prevotella. Finally, GH26 mannosidases and GH16 β-(1,3-1,4)-glucanase, mediating the breakdown of mannans and mixed glucans, also had similar bacterial origin, with several sequences affiliated with ruminal lineages, e.g., Acetovibrio, Fibrobacter, and Streptococcus.
Anaerobic fungi produce cellulosomes: extracellular structures that harbor multiple extracellular enzymes bound to scaffoldins (2). Cellulosome-bound genes in anaerobic fungi usually harbor a fungal dockerin domain (FDD) that is similar in structure to carbohydrate-binding module family 10 (CBM10) (5). We identified a total of 220 genes (see Table S21 in the supplemental material) with FDDs, 108 of which contained dual glycoside hydrolase-fungal dockerin domains (GH-FDDs). GH-FDD genes identified suggest that cellulosome-bound enzymes play a role in the degradation of cellulose and hemicellulose but not chitin, starch, or pectin. Within the remaining FDD-containing genes, we identified multiple putative activities that could either aid in biomass degradation (e.g., polysaccharide deacetylases, tannase, lipases, swollenin, and expansin module proteins) or act as cellulosomal preservation and defense mechanisms (e.g., protease inhibitors [serpins]), as well as multiple conserved hypothetical and hypothetical proteins.
Carbohydrate-binding modules (CBMs) are noncatalytic domains that are often encountered in lignocellulolytic enzymes and promote the association of the enzyme with the substrate. A total of 103 genes harboring carbohydrate-binding module (CBM) domains belonging to 6 different CBM families were identified (see Table S22 in the supplemental material). The majority (75.7%) of CBMs were members of the exclusively fungal CBM1 domain. Twenty-six genes with CBM domains were associated with GH enzymes, and 7 were associated with PL enzymes (see Table S22). Within GH-CBM dual domain genes, CBM1 domains were associated with several GH10 and GH11 xylanases, CBM18 domains were associated with GH18 chitinases, and CBM48 domains were associated with GH13 amylases. No CBM domains were identified in GH genes putatively involved in cellulose metabolism in the C1A genome. No CBM2 or CBM3 domains, the prevalent CBMs in bacterial plant biomass-degradation genes and in rumen anaerobic cellulosomal bacteria, respectively, were identified in the C1A genome (see Table S22).
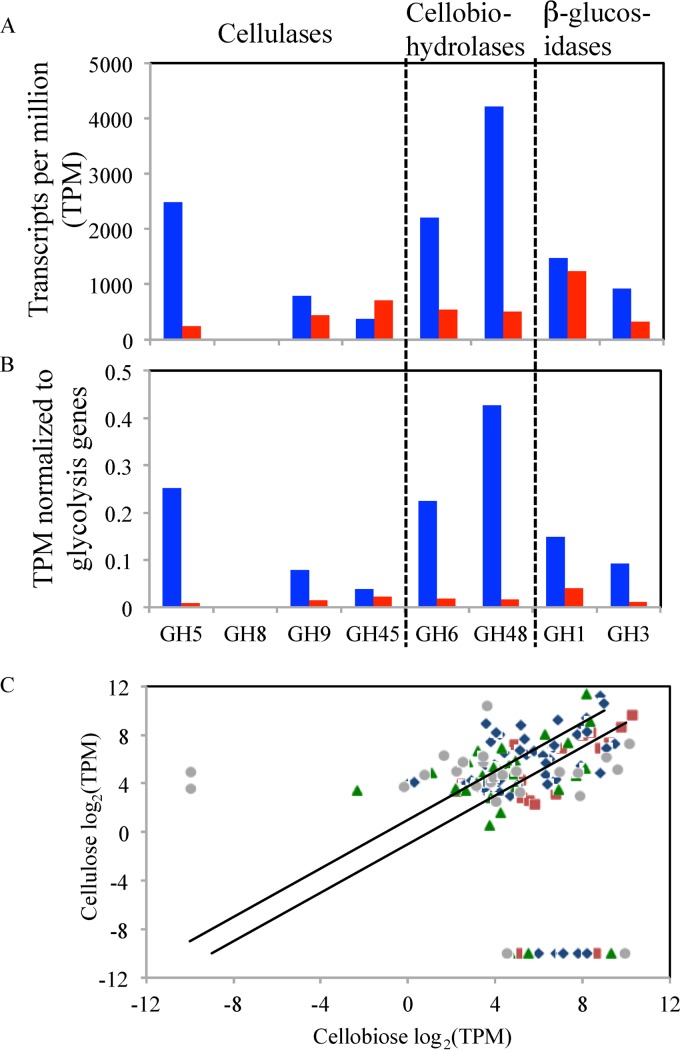
Comparative transcriptomic analysis of strain C1A was conducted on cellobiose-grown versus microcrystalline cellulose-grown cultures (Fig. 5; see also Fig. S6 and Tables S23 and S24 in the supplemental material). A total of 172 GH genes were expressed under both conditions, while 39 and 4 GH genes were identified only in cellobiose-grown and cellulose-grown cultures, respectively. In cellulose-grown cultures, transcripts belonging to GH5 cellulases, as well as GH9 and GH48 cellobiohydrolases, were drastically upregulated compared to cellobiose-grown cultures. GH8 and GH45 cellulases were only slightly upregulated, and their overall transcriptional levels were relatively low (Fig. 5A and B). GH1 and GH3 β-glucosidases, essential for substrate degradation under both conditions, were either not significantly changed or only slightly upregulated in cellulose-grown cultures (Fig. 5A and B).
Fig 5.
(A and B) Transcription levels of various GH family genes involved in cellulose degradation in cellulose (blue)- versus cellobiose (red)-grown cultures. Transcription levels are expressed as absolute transcripts per million (TPM) in panel A and as normalized TPM relative to a suite of glycolytic genes in panel B. (C) Differential GH gene expression by strain C1A grown on cellulose (y axis) and cellobiose (x axis) expressed as log2 TPM. Only genes with ≥10 TPM under at least one growth condition were used to construct the graph. The 2 diagonal lines represent boundaries between genes upregulated (above the upper line), downregulated (below the lower line), or not significantly changed (between the 2 lines) in cellulose- versus cellobiose-grown cultures. Color codes: blue, cellulases and cellobiohydrolases; red, β-glucosidases; green, other polymer-degrading GHs; gray, other oligomer-degrading GHs.
Analysis of expression profiles of all GH genes identified under both conditions revealed that while several cellulase and cellobiohydrolase genes were clearly upregulated in cellulose-grown cultures, the majority of such genes were not significantly (>2-fold) affected by the growth condition (Fig. 5C), and few were even significantly downregulated. Interestingly, few of the genes upregulated in cellulose-grown cultures belong to GH families associated with the degradation of plant polymers other than cellulose, e.g., GH10 and GH11 xylanases, GH18 chitinases, and GH26 mannosidases (see Fig. S6 in the supplemental material).
Strain C1A is an effective, versatile biomass degrader.
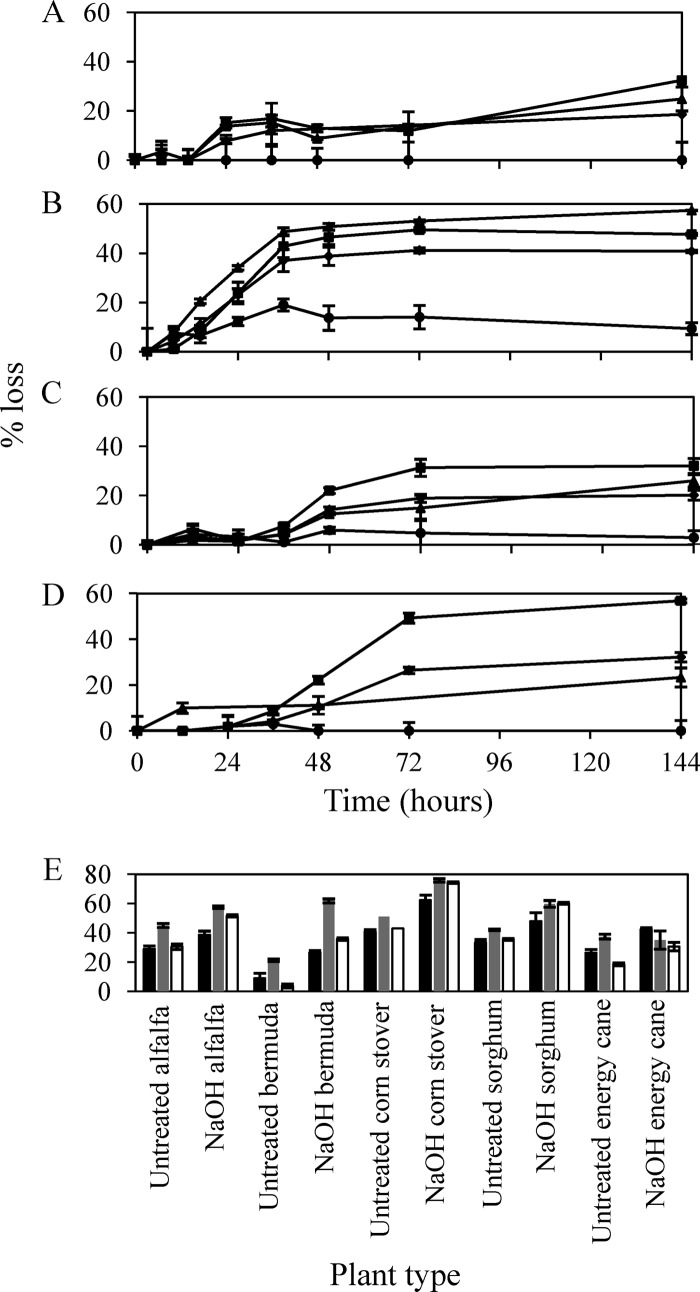
Strain C1A effectively metabolized a variety of sugars and polysaccharides, including crystalline cellulose and xylan (see Fig. S7 in the supplemental material). More importantly, strain C1A grew readily on untreated, as well as mild acid-, mild alkali-, and hydrothermolysis-treated switchgrass, with the concurrent utilization of cellulose and hemicellulose fractions, but not lignin (Fig. 6; also see Tables S25 and S26 and Fig. S8 and S9). Dry weight losses of substrate ranged from 18.6% (28.7% of nonlignin fraction) in untreated switchgrass to 40.8% (53.9% of nonlignin fraction) in NaOH-treated switchgrass. Further, adjustments to the inoculum/substrate ratios resulted in an increase in the amount of switchgrass metabolized up to 42.8% and 58.4% of the dry weight of untreated and NaOH-treated switchgrass, respectively. Strain C1A performed extremely well on NaOH-treated switchgrass, since this method of pretreatment retains the majority of the hemicellulose content (90, 91), which is degradable by strain C1A. Strain C1A also grew well on hydrothermolysis-treated switchgrass, presumably due to the fact that the removal of hemicellulose resulted in a greater accessibility to cellulose fibers. End product analysis indicated that lactate, acetate, and formate are the main end products of plant biomass degradation. Only minor amounts of ethanol were produced, ranging between 0.045 and 0.096 mg ethanol/mg biomass (Table S26).
Fig 6.
Lignocellulolytic capabilities of strain C1A. (A to D) Grams of dry weight (◆), cellulose (■), hemicellulose (▲), and lignin (●) lost in microcosms that contained untreated (A), sodium hydroxide-treated (B), acid-treated (C), and hydrothermolysis-treated (D) switchgrass. (E) Percentages of dry weight (black bars), cellulose (gray bars), and hemicellulose (white bars) lost in microcosms with different types of untreated and sodium hydroxide-treated plant materials.
In addition to switchgrass, we tested the capability of strain C1A to utilize several other types of energy crops (e.g., alfalfa, sorghum, and energy cane), agricultural residues (e.g., corn stover), and grasses (e.g., Bermuda grass). We chose these specific plant materials due to the variations in the percentages of cellulose, hemicellulose, and lignin in these plants. The results (Fig. 6E; see also Tables S25 and S26 in the supplemental material) demonstrate the versatility of strain C1A, since it was able to metabolize all different types of examined plant biomass. Within both untreated and NaOH-treated experiments, strain C1A was most effective in metabolism of corn stover, with 40.6% and 62.3% dry weight loss, 51.0% and 75.8% loss in cellulose fraction, and 43.0% and 74.3% loss in hemicellulose fractions in untreated and NaOH-treated corn stover, respectively.
DISCUSSION
Analysis of the C1A genome revealed thoroughly eukaryotic information processing, cytoskeletal structure, and intracellular trafficking machineries. On the other hand, we identified multiple cellular processes in which the C1A genome possesses features that appear to be absent from Dikarya genomes are but mostly associated with early-branching fungi and nonfungal Opisthokonta (Table 1). These observations suggest that such features have evolved prior to fungal separation from an Opisthokonta ancestor and were subsequently lost during the evolution of Dikarya but were retained in the Neocallimastigomycota. The rationale behind the retention of some of these features in the Neocallimastigomycota could be attributed to their unique habitat and evolutionary trajectory. For example, the possession of protease inhibitors to guard against plant, ciliate, and bacterial proteases is extremely beneficial in the rumen habitat. The possession of an axoneme and an intraflagellar-trafficking machinery is required for the motility of flagellated zoospores produced by the Neocallimastigomycota but not the Dikarya. However, the rationale behind other observed differences, e.g., retention of specific intramembrane proteases, posttranslational fucosylation capabilities, or the majority of focal adhesion proteins in the Neocallimastigomycota, is not entirely clear.
Many of the observed structural, metabolic, and genomic traits within the C1A genome are not shared with other early-branching fungal relatives or nonfungal Opisthokonta and hence could be regarded as Neocallimastigomycota-specific adaptations to the anaerobic gut environment. The mitochondrial reductive evolution to a hydrogenosome, the apparent replacement of ergosterol with tetrahymanol in the cell membrane (since oxygen is required for squalene epoxidation, steroid ring demethylation, and ring unsaturation during ergosterol biosynthesis [92]), and the sole dependence on a mixed-acid fermentation pathway for pyruvate metabolism and energy production in strain C1A are clear adaptations to anaerobiosis. The development of cellulosomes and the acquisition of many GH enzymes could be viewed as an adaptation to improve the access, speed, and efficacy of biomass degradation.
In addition to metabolic adaptations to an O2-independent mode of metabolism and organelle development via reductive evolution and gene acquisition, evolution of anaerobic fungi in the rumen and gut of herbivores appears to have triggered multiple genome-wide patterns. These include the possession of a large genome, the presence of large intergenic regions, the low (17.0%) G+C content, and the occurrence of a high level of gene duplication and microsatellite repeats (Fig. 1; see also Text S1 and Tables S4, S7, and S8 in the supplemental material). We argue that these genome-wide patterns are due to genetic drift, triggered by the low effective population sizes, bottlenecks in vertical transmission, and the asexual life style of anaerobic fungi. Species with low effective population sizes could tolerate slightly deleterious accumulation of DNA, resulting in the expansion in genome size, accumulation of repeats, and gene duplications (93, 94). In addition, genetic drift is also associated with an increase in the rate of nonlethal mutations, which tend to be biased toward adenine or thymine mutations such as cytosine deamination or guanine oxidation (95).
This study also highlights the extensive lignocellulolytic machinery and robust plant biomass degradation capability of strain C1A, observations which are consistent with prior studies identifying multiple cellulolytic and hemicellulolytic genes from anaerobic fungal strains (5, 14, 15, 66–88) and documenting the capability of such strains to degrade various plant substrates (96–99). Further, this study clearly demonstrates that the GH machinery in the C1A genome is markedly different from that of aerobic lignocellulolytic fungi. Such differences appear to be driven by physiological considerations, variations in the employed biomass degradation strategy, and habitat distinction. The recent demonstration of an O2-dependent mode of metabolism for GH61 enzymes could explain the ubiquity of this family in aerobic fungal genomes and its absence in the C1A genome (100). The utilization of a cellulosomal strategy for plant biomass degradation by strain C1A, compared to the free extracellular enzyme strategy of aerobic fungi, could explain the identification of a large number of GH genes with a dockerin domain in the C1A genome. Finally, the rumen habitat of the Neocallimastigomycota and the widespread gene acquisition of bacterial GH genes in the C1A genome could explain the occurrence of GH genes belonging to families rarely encountered in aerobic fungi, e.g., GH8 and GH48.
Interestingly, while gene acquisition from prevalent rumen bacterial lineages plays an important role in shaping the C1A lignocellulolytic machinery, a fraction of C1A GH genes were associated with bacterial lineages that are not regarded as integral members of the bovine rumen microbiota, e.g., the taxa Actinobacteria, Thermotoga, Deinococcus, and Chloroflexi. This intriguing observation could possibly be explained by the occasional identification of some of these taxa as minor components in the bovine rumen (101). Further, the extensively studied bovine rumen should not be regarded as the only possible habitat for anaerobic fungal gene acquisition, since anaerobic fungi have a wide distribution in the rumen, hindgut, and feces of multiple ruminant and nonruminant herbivores (2). Finally, it is important to note that evolution of anaerobic fungi from an Opisthokonta ancestor has preceded the evolution of their metazoan herbivore hosts (102). As such, anaerobic fungi could have acquired such genes prior to their association with the reptilian or mammalian alimentary tracts.
Transcriptional studies indicated that a large number of polymer-degrading GH genes are constitutively expressed in cellobiose-grown cultures. However, C1A cellulose-grown cultures exhibited significant increases in the overall transcription levels of specific cellulase (GH5) and cellobiohydrolase (GH9 and GH48) GH families, suggesting a prominent role for these three families in cellulose metabolism. The increase in overall levels of transcripts belonging to a specific GH family was mainly attributed to the upregulation of a fraction of its genes (Fig. 5C). For example, while the overall transcriptional level of GH48 cellobiohydrolases increased 8-fold in cellulose-grown cultures, only 5 out of 12 genes were upregulated in cellulose-grown cultures, while 2 genes were not significantly impacted and 4 were downregulated. Factors influencing this observed selective regulation remain to be elucidated.
Finally, our results suggest that the lignocellulolytic capabilities of strain C1A could be exploited outside the rumen for the production of biofuels from plant biomass. The most promising approach for lignocellulosic biofuel production involves consolidated bioprocessing, which combines the saccharification of lignocellulose and the fermentation of the resulting sugars in a single step and is carried out by a single microorganism or microbial consortium (103). Here, we show that strain C1A simultaneously couples the saccharification of the cellulosic and hemicellulosic fractions of plants to the fermentation of the resulting hexose and pentose sugars. Further, the invasive nature and filamentous growth pattern of these anaerobic fungi allow plant biomass degradation to proceed without pretreatment, and the process was significantly enhanced using mild pretreatments (Fig. 6). To our knowledge, the extent of lignocellulosic biomass degradation by strain C1A has not been reported for a single microorganism in the absence of saccharification enzymes. Anaerobic fungi thus represent extremely promising microorganisms for exploitation in direct lignocellulolytic schemes. As part of its fermentative metabolism, strain C1A is capable of producing ethanol as a minor end product during pyruvate metabolism. Indeed, 1 copy of alcohol dehydrogenase has been identified, and C1A can tolerate up to 3% ethanol (data not shown). However, given its relatively low ethanol productivity and relatively low ethanol tolerance, efforts toward improving alcohol production and tolerance via physiological and genetic manipulations are needed to improve ethanol productivity in this remarkable plant biomass-degrading anaerobic fungal strain.
Supplementary Material
ACKNOWLEDGMENTS
We thank Yanqi Wu and Gopal Kakani for providing plant materials and Tammy Austin for editorial assistance.
This work was supported by NSF EPSCoR award EPS 0814361 and utilized computational capacities supported by NSF grant number OCI-1053575.
Footnotes
Published ahead of print 24 May 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00821-13.
REFERENCES
- 1. Orpin CG. 1975. Studies on the rumen flagellate Neocallimastix frontalis. J. Gen. Microbiol. 91:249–262 [DOI] [PubMed] [Google Scholar]
- 2. Orpin CG. 1994. Anaerobic fungi: taxonomy, biology, and distribution in nature, p 1–45 Mountfort DO, Orpin CG. (ed), Anaerobic fungi: biology, ecology, and function. Marcel Dekker, Inc, New York, NY [Google Scholar]
- 3. Ma L-J, Ibrahim AS, Skory C, Grabherr MG, Burger G, Butler M, Elias M, Idnurm A, Lang BF, Sone T, Abe A, Calvo SE, Corrochano LM, Engels R, Fu J, Hansberg W, Kim J-M, Kodira CD, Koehrsen MJ, Liu B, Miranda-Saavedra D, O'Leary S, Ortiz-Castellanos L, Poulter R, Rodriguez-Romero J, Ruiz-Herrera J, Shen Y-Q, Zeng Q, Galagan J, Birren BW, Cuomo CA, Wickes BL. 2009. Genomic analysis of the basal lineage fungus Rhizopus oryzae reveals a whole-genome duplication. PLoS Genet. 5:e1000549. 10.1371/journal.pgen.1000549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Joneson S, Stajich JE, Shiu S-H, Rosenblum EB. 2011. Genomic transition to pathogenicity in chytrid fungi. PLoS Pathog. 7:e1002338. 10.1371/journal.ppat.1002338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ljungdahl LG. 2008. The cellulase/hemicellulase system of the anaerobic fungus Orpinomyces PC-2 and aspects of its use. Ann. N. Y. Acad. Sci. 1125:308–321 [DOI] [PubMed] [Google Scholar]
- 6. Mentel M, Martin W. 2008. Energy metabolism among eukaryotic anaerobes in light of Proterozoic ocean chemistry. Philos. Trans. R. Soc. B 363:2717–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schneider RE, Brown MT, Shiflett AM, Dyall SD, Hayes RD, Xie Y, Loo JA, Johnson PJ. 2011. The Trichomonas vaginalis hydrogenosome proteome is highly reduced relative to mitochondria, yet complex compared with mitosomes. Int. J. Parasitol. 41:1421–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Müller M, Mentel M, van Hellemond JJ, Henze K, Woehle C, Gould SB, Yu R-Y, van der Giezen M, Tielens AGM, Martin WF. 2012. Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol. Mol. Biol. Rev. 76:444–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tachezy J, Doležal P. 2007. Iron-sulfur proteins and iron-sulfur cluster assembly in organisms with hydrogenosomes and mitosomes, p 105–134 Martin WF, M̈uller M. (ed), Origin of mitochondria and hydrogenosomes. Springer-Verlag, Berlin, Germany [Google Scholar]
- 10. Lowe SE, Griffith GG, Milne A, Theodorou MK, Trinci APJ. 1987. The life cycle and growth kinetics of an anaerobic rumen fungus. J. Gen. Microbiol. 133:1815–1827 [Google Scholar]
- 11. Hibbett DS, Binder M, Bischoff JF, Blackwell M, Cannon PE, Eriksson OE, Huhndorf S, James T, Kirk PM, Lucking R, Thorsten LH, Lutzoni F, Matheny PB, McLaughlin DJ, Powell MJ, Redhead S, Schoch C, Spatafora JW, Stalpers JA, Vilgalys R, Aime MC, Aptroot A, Bauer R, Begerow D, Benny GL, Castlebury LA, Crous PW, Dai Y-C, Gams W, Geiser D, Griffith GW, Gueidan C, Hawksworth DL, Hestmark G, Hosaka K, Humber RA, Hyde KD, Ironside JE, Koljalg U, Kurtzman CP, Larsson K-H, Lichtwardt R, Longcore J, Miadlikowska J, Miller A, Moncalvo J-M, Mozley-Standridge S, Oberwinkler F, Parmasto EE, Reeb VV, Rogers JD, Roux C, Ryvarden L, Sampaio J, Schussler AF, Sugiyama J, Thorn RG, Tibell L, Untereiner WA, Walker C, Wang Z, Weir A, Weiss M, White M, Winka K, Yao Y-J, Zhang N. 2007. A higher-level phylogenetic classification of the Fungi. Mycol. Res. 111:509–547 [DOI] [PubMed] [Google Scholar]
- 12. Lowe SE, Theodorou M, Trinci A. 1987. Growth and fermentation of an anaerobic rumen fungus on various carbon sources and effect of temperature on development. Appl. Environ. Microbiol. 53:1210–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Raghothama S, Eberhardt RY, Simpson P, Wigelsworth D, White P, Hazlewood GP, Nagy T, Gilbert HJ, Williamson MP. 2001. Characterization of a cellulome dockerin domain from the anaerobic fungus Piromyces equi. Nat. Struct. Biol. 8:775–778 [DOI] [PubMed] [Google Scholar]
- 14. Williams AG, Orpin CG. 1987. Glycoside hydrolase enzymes present in the zoospore and vegetative growth stages of the rumen fungi Neocallimastix patriciarum, Piromonas communis, and an unidentified isolate, grown on a range of carbohydrates. Can. J. Microbiol. 33:427–434 [DOI] [PubMed] [Google Scholar]
- 15. Williams AG, Orpin CG. 1987. Polysaccharide-degrading enzymes formed by three species of anaerobic rumen fungi grown on a range of carbohydrate substrates. Can. J. Microbiol. 33:418–426 [DOI] [PubMed] [Google Scholar]
- 16. Theodorou MK, Brookman J, Trinci A. 2005. Anaerobic fungi, p 55–66 Makkar HP, McSweeney CS. (ed), Methods in gut microbial ecology for ruminants. Springer, Dordrecht, The Netherlands [Google Scholar]
- 17. Marvin-Sikkema F, Richardson A, Stewart C, Gottschal J, Prins R. 1990. Influence of hydrogen-consuming bacteria on cellulose degradation by anaerobic fungi. Appl. Environ. Microbiol. 56:3793–3797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bryant M. 1972. Commentary on the Hungate technique for culture of anaerobic bacteria. Am. J. Clin. Nutr. 25:1324–1328 [DOI] [PubMed] [Google Scholar]
- 19. Balch WE, Wolfe R. 1976. New approach to the cultivation of methanogenic bacteria: 2-mercaptoethanesulfonic acid (HS-CoM)-dependent growth of Methanobacterium ruminantium in a pressureized atmosphere. Appl. Environ. Microbiol. 32:781–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brownlee AG. 1989. Remarkably AT-rich genomic DNA from the anaerobic fungus Neocallimastix. Nucleic Acids Res. 17:1327–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, Boutell JM, Bryant J, Carter RJ, Cheetham RK, Cox AJ, Ellis DJ, Flatbush MR, Gormley NA, Humphray SJ, Irving LJ, Karbelashvili MS, Kirk SM, Li H, Liu X, Maisinger KS, Murray LJ, Obradovic B, Ost T, Parkinson ML, Pratt MR, Rasolonjatovo IM, Reed MT, Rigatti R, Rodighiero C, Ross MT, Sabot A, Sankar SV, Scally A, Schroth GP, Smith ME, Smith VP, Spiridou A, Torrance PE, Tzonev SS, Vermaas EH, Walter K, Wu X, Zhang L, et al. 2008. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456:53–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, Bibillo A, Bjornson B, Chaudhuri B, Christians F, Cicero R, Clark S, Dalal R, Dewinter A, Dixon J, Foquet M, Gaertner A, Hardenbol P, Heiner C, Hester K, Holden D, Kearns G, Kong K, Kuse R, Lacroix Y, Lin S, Lundquist P, Ma C, Marks P, Maxham M, Murphy D, Park I, Pham T, Phillips M, Roy J, Sebra R, Shen G, Sorenson J, Tomaney A, Travers K, Trulson M, Vieceli J, Wegener J, Wu D, Yang A, Zaccarin D, Zhao P, Zhong F, Korlach J, Turner S. 2009. Real-time DNA sequencing from single polymerase molecules. Science 323 133–138 [DOI] [PubMed] [Google Scholar]
- 23. Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Koren S, Schatz MC, Walenz BP, Martin J, Howard JT, Ganapathy G, Wang Z, Rasko DA, McCombie WR, Jarvis ED, Phillippy AM. 2012. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat. Biotechnol. 30:693–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Myers EW, Sutton GG, Delcher AL, Dew IM, Fasulo DP, Flanigan ML, Kravitz SA, Mobarry CM, Reinert KH, Remington KA, Anson EL, Bolanos RA, Chou HH, Jordan CM, Halpern AL, Lonardi S, Beasley EM, Brandon RC, Chen L, Dunn PJ, Lai Z, Liang Y, Nusskern DR, Zhan M, Zhang Q, Zheng X, Rubin GM, Adams MD, Venter JC. 2000. A whole-genome assembly of Drosophila. Science 287:2196–2204 [DOI] [PubMed] [Google Scholar]
- 26. Miller JR, Delcher AL, Kore S, Venter E, Walenz BP, Brownley A, Johnson J, Li K, Mobarry C, Sutton G. 2008. Aggressive assembly of pyrosequencing reads with mates. Bioinformatics 24:2818–2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Magoc T, Salzberg SL. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ewing B, Hillier L, Wendl MC, Green P. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175–185 [DOI] [PubMed] [Google Scholar]
- 29. Parra G, Bradnam K, Korf I. 2007. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23:1061–1067 [DOI] [PubMed] [Google Scholar]
- 30. Wang Z, Gerstein M, Snyder M. 2009. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10:57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29:644–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Majoros WH, Pertea M, Salzberg SL. 2004. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics 20:2878–2879 [DOI] [PubMed] [Google Scholar]
- 34. Stanke M, Schöffmann O, Morgenstern B, Waack S. 2006. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics 7:62. 10.1186/1471-2105-7-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Haas BJ, Delcher AL, Mount SM, Wortman JR, Smith RK, Jr, Hannick LI, Maiti R, Ronning CM, Rusch DB, Town CD, Salzberg SL, White O. 2003. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 31:5654–5666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu TD, Watanabe CK. 2005. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21:1859–1875 [DOI] [PubMed] [Google Scholar]
- 37. Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput. Biol. 10:e1002195. 10.1371/journal.pcbi.1002195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer EL, Eddy SR, Bateman A, Finn RD. 2012. The Pfam protein families database. Nucleic Acids Res. 40:D290–D301. 10.1093/nar/gkr1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hess M, Sczyrba A, Egan R, Kim TW, Chokhawala H, Schroth G, Luo S, Clark DS, Chen F, Zhang T, Mackie RI, Pennacchio LA, Tringe SG, Visel A, Woyke T, Wang Z, Rubin EM. 2011. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science 331:463–467 [DOI] [PubMed] [Google Scholar]
- 41. Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. 2009. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37:D233–D238. 10.1093/nar/gkn663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8:785–786 [DOI] [PubMed] [Google Scholar]
- 43. Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580 [DOI] [PubMed] [Google Scholar]
- 44. Jurka J, Bao W, Kojima K, Kapitonov VV. 15 February 2011. Repetitive elements: bioinformatic identification, classification and analysis. eLS 10.1002/9780470015902.a0005270.pub2 [DOI] [Google Scholar]
- 45. Dhillon B, Goodwin SB. 2011. Identification and annotation of repetitive sequences in fungal genomes. Methods Mol. Biol. 722:33–50 [DOI] [PubMed] [Google Scholar]
- 46. Floudas D, Binder M, Riley R, Barry K, Blanchette RA, Henrissat B, Martínez AT, Otillar R, Spatafora JW, Yadav JS, Aerts A, Benoit I, Boyd A, Carlson A, Copeland A, Coutinho PM, de Vries RP, Ferreira P, Findley K, Foster B, Gaskell J, Glotzer D, Górecki P, Heitman J, Hesse C, Hori C, Igarashi K, Jurgens JA, Kallen N, Kersten P, Kohler A, Kües U, Kumar TKA, Kuo A, LaButti K, Larrondo LF, Lindquist E, Ling A, Lombard V, Lucas S, Lundell T, Martin R, McLaughlin DJ, Morgenstern I, Morin E, Murat C, Nagy LG, Nolan M, Ohm RA, et al. 2012. The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 336:1715–1719 [DOI] [PubMed] [Google Scholar]
- 47. Price AL, Jones NC, Pevzner PA. 2005. De novo identification of repeat families in large genomes. Bioinformatics 21:i351–i358 [DOI] [PubMed] [Google Scholar]
- 48. Xu Z, Wang H. 2007. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 35:W265–W268. 10.1093/nar/gkm286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689. 10.1093/nar/gki366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Claros MG, Vincens P. 1996. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 241:779–786 [DOI] [PubMed] [Google Scholar]
- 51. Thomas PD, Campbell MJ, Kejariwal A, Huaiyu M, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A. 2003. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 13:2129–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Suryawati L, Wilkins WR, Bellmer DD, Huhnke RL, Maness NO, Banat IM. 2008. Simultaneous saccharification and fermentation of Kanlow switchgrass pretreated by hydrothermolysis using Kluyveromyces marxianus IMB4. Biotechnol. Bioeng. 101:894–902 [DOI] [PubMed] [Google Scholar]
- 53. Xu J, Cheng JJ, Sharma-Shivappa RR, Burns JC. 2010. Sodium hydroxide pretreatment of switchgrass for ethanol production. Energy Fuels 24:2113–2119 [Google Scholar]
- 54. Torget R, Werdene P, Himmel M, Grohmann K. 1990. Dilute acid pretreatment of short rotation woody and herbaceous crops. Appl. Biochem. Biotechnol. 24:115–126 [Google Scholar]
- 55. Sluiter A, Hames B, Ruiz R, Scarlata C, Sluiter J, Templeton D, Crocker D. 2008. Determination of structural carbohydrates and lignin in biomass. Technical report NREL/TP-510-42618 National Renewable Energy Laboratory, Golden, CO [Google Scholar]
- 56. Sluiter J, Sluiter A. 2010. Summative mass closure. Technical report NREL/TP-510-48825 National Renewable Energy Laboratory, Golden, CO [Google Scholar]
- 57. Faga BA, Wilkins RM, Banat IM. 2010. Ethanol production through simultaneous saccharification and fermentation of switchgrass using Saccharomyces cerevisiae D5A and thermotolerant Kluyveromyces marxianus IMB strains. Bioresour. Technol. 101:2273–2279 [DOI] [PubMed] [Google Scholar]
- 58. Karaoglu H, Lee CMY, Meyer W. 2005. Survey of simple sequence repeats in completed fungal genomes. Mol. Biol. Evol. 22:639–649 [DOI] [PubMed] [Google Scholar]
- 59. Sebé-Pedrós A, Roger AJ, Lang FB, King N, Ruiz-Trillo I. 2010. Ancient origin of the integrin-mediated adhesion and signaling machinery. Proc. Natl. Acad. Sci. U. S. A. 107:10142–10147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Steenbakkers PJM, Irving JA, Harhangi HR, Swinkels WJC, Akhmanova A, Dijkerman R, Jetten MSM, van der Drift C, Whisstock JC, Op den Camp HJM. 2008. A serpin in the cellulosome of the anaerobic fungus Piromyces sp. strain E2. Mycol. Res. 112:999–1006 [DOI] [PubMed] [Google Scholar]
- 61. Rada P, Doležal P, Jedelský PL, Bursac D, Perry AJ, Šedinová M, Smíšková K, Novotný M, Beltrán NC, Hrdý I, Lithgow T, Tachezy J. 2011. The core components of organelle biogenesis and membrane transport in the hydrogenosomes of Trichomonas vaginalis. PLoS One 6:e24428. 10.1371/journal.pone.0024428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Boxma B, Voncken F, Jannink S, Van Alen T, Akhmanova A, Van Weelden SWH, Van Hellemond JJ, Ricard G, Huynen M, Tielens AGM, Hackstein JHP. 2004. The anaerobic chytridiomycete fungus Piromyces sp. E2 produces ethanol via pyruvate:formate lyase and an alcohol dehydrogenase E. Mol. Microbiol. 51:1389–1399 [DOI] [PubMed] [Google Scholar]
- 63. Yarlett N, Orpin CG, Munn EA, Yarlett NC, Greenwood CA. 1986. Hydrogenosomes in the rumen fungus Neocallimastix patriciarum. Biochem. J. 236:729–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hackstein JHP, Baker SE, van Hellemond JJ, Tielens AGM. 2008. Hydrogenosomes of anaerobic chytrids: an alternative way to adapt to anaerobic environments, p 147–162 Tachezy J. (ed), Hydrogenosomes and mitosomes: mitochondria of anaerobic eukaryotes. Springer-Verlag, Berlin, Germany [Google Scholar]
- 65. Hackstein J, Tjaden J, Koopman WJH, Huynen M. 2007. Hydrogenosomes (and related organelles, either) are not the same, p 135–160 Martin WF, M̈uller M. (ed), Origin of mitochondria and hydrogenosomes. Springer Verlag, New York, NY [Google Scholar]
- 66. Chen H, Hopper S, Li X-L, Ljungdahl L, Cerniglia C. 2006. Isolation of extremely AT-rich genomic DNA and analysis of genes encoding carbohydrate-degrading enzymes from Orpinomyces sp. strain PC-2. Curr. Microbiol. 53:396–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chen H, Li X-L, Blum DL, Ximenes E, Ljungdahl L. 2003. CelF of Orpinomyces PC-2 has an intron and encodes a cellulase (CelF) containing a carbohydrate-binding module. Appl. Biochem. Biotechnol. 105-108:775–785 [DOI] [PubMed] [Google Scholar]
- 68. Chen H, Li XL, Ljungdahl LG. 1997. Sequencing of a 1,3-1,4-beta-D-glucanase (lichenase) from the anaerobic fungus Orpinomyces strain PC-2: properties of the enzyme expressed in Escherichia coli and evidence that the gene has a bacterial origin. J. Bacteriol. 179:6028–6034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chen H-L, Chen Y-C, Lu M-Y, Chang J-J, Wang H-T, Ke H-M, Wang T-Y, Ruan S-K, Wang T-Y, Hung K-Y, Cho H-Y, Lin W-T, Shih M-C, Li W-H. 2012. A highly efficient beta-glucosidase from the buffalo rumen fungus Neocallimastix patriciarum W5. Biotechnol. Biofuels 5:24. 10.1186/1754-6834-5-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Durand R, Rascle C, Fèvre M. 1996. Molecular characterization of xyn3, a member of the endoxylanase multigene family of the rumen anaerobic fungus Neocallimastix frontalis. Curr. Genet. 30:531–540 [DOI] [PubMed] [Google Scholar]
- 71. Eberhardt RY, Gilbert HJ, Hazlewood GP. 2000. Primary sequence and enzymic properties of two modular endoglucanases, Cel5A and Cel45A, from the anaerobic fungus Piromyces equi. Microbiology 146:1999–2008 [DOI] [PubMed] [Google Scholar]
- 72. Fanutti C, Ponyi T, Black GW, Hazlewood GP, Gilbert HJ. 1995. The conserved noncatalytic 40-residue sequence in cellulases and hemicellulases from anaerobic fungi functions as a protein docking domain. J. Biol. Chem. 270:29314–29322 [DOI] [PubMed] [Google Scholar]
- 73. Fujino Y, Ogata K, Nagamine T, Ushida K. 1998. Cloning, sequencing, and expression of an endoglucanase gene from the rumen anaerobic fungus Neocallimastix frontalis MCH3. Biosci. Biotechnol. Biochem. 62:1795–1798 [DOI] [PubMed] [Google Scholar]
- 74. Harhangi HR, Akhmanova A, Steenbakkers PJM, Jetten MSM, van der Drift C, Op den Camp HJM. 2003. Genomic DNA analysis of genes encoding (hemi-)cellulolytic enzymes of the anaerobic fungus Piromyces sp. E2. Gene 314:73–80 [DOI] [PubMed] [Google Scholar]
- 75. Harhangi HR, Akhmanova AS, Emmens R, van der Drift C, de Laat WTAM, van Dijken JP, Jetten MSM, Pronk JT, Op den Camp HJM. 2003. Xylose metabolism in the anaerobic fungus Piromyces sp. strain E2 follows the bacterial pathway. Arch. Microbiol. 180:134–141 [DOI] [PubMed] [Google Scholar]
- 76. Harhangi HR, Freelove ACJ, Ubhayasekera W, van Dinther M, Steenbakkers PJM, Akhmanova A, van der Drift C, Jetten MSM, Mowbray SL, Gilbert HJ, Op den Camp HJM. 2003. Cel6A, a major exoglucanase from the cellulosome of the anaerobic fungi Piromyces sp. E2 and Piromyces equi. Biochim. Biophys. Acta 1628:30–39 [DOI] [PubMed] [Google Scholar]
- 77. Harhangi HR, Steenbakkers PJM, Akhmanova A, Jetten MSM, van der Drift C, Op den Camp HJM. 2002. A highly expressed family 1 β-glucosidase with transglycosylation capacity from the anaerobic fungus Piromyces sp. E2. Biochim. Biophys. Acta 1574:293–303 [DOI] [PubMed] [Google Scholar]
- 78. Li X-L, Ljungdahl L, Ximenes E, Chen H, Felix C, Cotta M, Dien B. 2004. Properties of a recombinant β-glucosidase from polycentric anaerobic fungus Orpinomyces PC-2 and its application for cellulose hydrolysis. Appl. Biochem. Biotechnol. 113:233–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Liu J-H, Selinger BL, Tsai C-F, Cheng K-J. 1999. Characterization of a Neocallimastix patriciarum xylanase gene and its product. Can. J. Microbiol. 45:970–974 [DOI] [PubMed] [Google Scholar]
- 80. Madhavan A, Tamalampudi S, Ushida K, Kanai D, Katahira S, Srivastava A, Fukuda H, Bisaria V, Kondo A. 2009. Xylose isomerase from polycentric fungus Orpinomyces gene sequencing, cloning, and expression in Saccharomyces cerevisiae for bioconversion of xylose to ethanol. Appl. Microbiol. Biotechnol. 82:1067–1078 [DOI] [PubMed] [Google Scholar]
- 81. Nicholson MJ, Theodorou MK, Brookman JL. 2005. Molecular analysis of the anaerobic rumen fungus Orpinomyces—insights into an AT-rich genome. Microbiology 151:121–133 [DOI] [PubMed] [Google Scholar]
- 82. Pai C-K, Wu Z-Y, Chen M-J, Zeng Y-F, Chen J-W, Duan C-H, Li M-L, Liu J-R. 2010. Molecular cloning and characterization of a bifunctional xylanolytic enzyme from Neocallimastix patriciarum. Appl. Microbiol. Biotechnol. 85:1451–1462 [DOI] [PubMed] [Google Scholar]
- 83. Qiu X, Selinger B, Yanke LJ, Cheng KJ. 2000. Isolation and analysis of two cellulase cDNAs from Orpinomyces joyonii. Gene 245:119–126 [DOI] [PubMed] [Google Scholar]
- 84. Steenbakkers P, Freelove A, Van Cranenbroek B, Sweegers B, Harhangi H, Vogels G, Hazlewood G, Gilbert H, Op den Camp H. 2002. The major component of the cellulosomes of anaerobic fungi from the genus Piromyces is a family 48 glycoside hydrolase. DNA Seq. 13:313–320 [DOI] [PubMed] [Google Scholar]
- 85. Steenbakkers PJM, Harhangi HR, Bosscher MW, van der Hooft MMC, Keltjens JT, van der Drift C, Vogels GD, Op den Camp HJM. 2003. Beta-glucosidase in cellulosome of the anaerobic fungus Piromyces sp. strain E2 is a family 3 glycoside hydrolase. Biochem. J. 370:963–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Steenbakkers PJM, Ubhayasekera W, Goossen HJAM, van Lierop EMHM, van der Drift C, Vogels GD, Mowbray SL, Op den Camp HJM. 2002. An intron-containing glycoside hydrolase family 9 cellulase gene encodes the dominant 90 kDa component of the cellulosome of the anaerobic fungus Piromyces sp. strain E2. Biochem. J. 365:193–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Xue G-P, Gobius KS, Orpin CG. 1992. A novel polysaccharide hydrolase cDNA (celD) from Neocallimastix patriciarum encoding three multi-functional catalytic domains with high endoglucanase, cellobiohydrolase and xylanase activities. J. Gen. Microbiol. 138:2397–2403 [DOI] [PubMed] [Google Scholar]
- 88. Zhou L, Xue GP, Orpin CG, Black GW, Gilbert HJ, Hazlewood GP. 1994. Intronless celB from the anaerobic fungus Neocallimastix patriciarum encodes a modular family A endoglucanase. Biochem. J. 297:359–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Scheller HV, Ulvskov P. 2010. Hemicelluloses. Annu. Rev. Plant Biol. 61:263–289 [DOI] [PubMed] [Google Scholar]
- 90. Alvira P, Thomas-Pejo E, Ballesteros M, Negro MJ. 2010. Pretreatment technologies for an efficient bioethanol production process based on enzymatic hydrolysis: a review. Bioresour. Technol. 101:4851–4861 [DOI] [PubMed] [Google Scholar]
- 91. Mosiera N, Wyman C, Dale B, Elander R, Le YY, Holtzapple M, Ladisch M. 2005. Features of promising technologies for pretreatment of lignocellulosic biomass. Bioresour. Technol. 96:673–686 [DOI] [PubMed] [Google Scholar]
- 92. Waldbauer JR, Newman DK, Summons RE. 2011. Microaerobic steroid biosynthesis and the molecular fossil record of Archean life. Proc. Natl. Acad. Sci. U. S. A. 108:13409–13414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Lynch M, Conery JS. 2003. The origins of genome complexity. Science 302:1401–1404 [DOI] [PubMed] [Google Scholar]
- 94. Kelkar YD, Ochman H. 2012. Causes and consequences of genome expansion in Fungi. Genome Biol. Evol. 4:13–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. McCutcheon JP, Moran NA. 2012. Extreme genome reduction in symbiotic bacteria. Nat. Rev. Microbiol. 13:13–26 [DOI] [PubMed] [Google Scholar]
- 96. Sijtsma L, Tan B. 1996. Degradation of perennial ryegrass leaf and stem cell walls by the anaerobic fungus Neocallimastix sp. strain CS3b. Appl. Environ. Microbiol. 62:1437–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Edwards JE, Kingston-Smith AH, Jimenez HR, Huws SA, Skot KP, Griffith GW, McEwan NR, Theodorou MK. 2008. Dynamics of initial colonization of nonconserved perennial ryegrass by anaerobic fungi in the bovine rumen. FEMS Microbiol. Ecol. 66:537–545 [DOI] [PubMed] [Google Scholar]
- 98. McSweeney CS, Dulieu A, Katayama Y, Lowry JB. 1994. Solubilization of lignin by the ruminal anaerobic fungus Neocallimastix patriciarum. Appl. Environ. Microbiol. 60:2985–2989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Borneman W, Hartley R, Morrison WH, Akin D, Ljungdahl L. 1990. Feruloyl and p-coumaroyl esterase from anaerobic fungi in relation to plant cell wall degradation. Appl. Microbiol. Biotechnol. 33:345–351 [Google Scholar]
- 100. Quinlan RJ, Sweeney MD, Lo Leggio L, Otten H, Poulsen J-C, Johansen KS, Krogh KB, Jørgensen CI, Tovborg M, Anthonsen A, Tryfona T, Walter CP, Dupree P, Xu F, Davies GJ, Walton PH. 2011. Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. Proc. Natl. Acad. Sci. U. S. A. 108:15079–15084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Kim M, Morrison M, Yu Z. 2011. Status of the phylogenetic diversity census of ruminal microbiomes. FEMS Microbiol. Ecol. 76:49–63 [DOI] [PubMed] [Google Scholar]
- 102. Lücking R, Huhndorf S, Pfister DH, Plata ER, Lumbsch HT. 2009. Fungi evolved right on track. Mycologia 101:810–822 [DOI] [PubMed] [Google Scholar]
- 103. Xu Q, Singh A, Himmel ME. 2009. Perspectives and new directions for the production of bioethanol using consolidated bioprocessing of lignocellulose. Curr. Opin. Biotechnol. 20:364–371 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.