Pyruvate dehydrogenase complex

Pyruvate dehydrogenase complex (PDC) is a complex of three enzymes that converts pyruvate into acetyl-CoA by a process called pyruvate decarboxylation.[1] Acetyl-CoA may then be used in the citric acid cycle to carry out cellular respiration, and this complex links the glycolysis metabolic pathway to the citric acid cycle. Pyruvate decarboxylation is also known as the "pyruvate dehydrogenase reaction" because it also involves the oxidation of pyruvate.[2]
This multi-enzyme complex is related structurally and functionally to the oxoglutarate dehydrogenase and branched-chain oxo-acid dehydrogenase multi-enzyme complexes.
Reaction
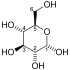
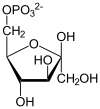
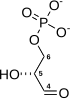
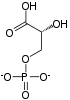
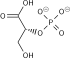
The reaction catalysed by pyruvate dehydrogenase complex is:
| pyruvate | pyruvate dehydrogenase complex | acetyl CoA | |

|
|||
| CoA-SH + NAD+ | CO2 + NADH + H+ | ||

| |||
Structure
Pyruvate dehydrogenase (E1)

The E1 subunit, called the pyruvate dehydrogenase subunit, is either a homodimer (comprising two “α” chains, e.g. in Escherichia coli) or a heterotetramer of two different chains (two “α” and two “β” chains). A magnesium ion forms a 4-coordinate complex with three, polar amino acid residues (Asp, Asn, and Tyr) located on the alpha chain, and the thiamine diphosphate (TPP) cofactor directly involved in decarboxylation of the pyruvate.[3][4]
Dihydrolipoyl transacetylase (E2)
The E2 subunit, or dihydrolipoyl acetyltransferase, for both prokaryotes and eukaryotes, is generally composed of three domains. The N-terminal domain (the lipoyl domain), consists of 1–3 lipoyl groups of approximately 80 amino acids each. The peripheral subunit binding domain (PSBD), serves as a selective binding site for other domains of the E1 and E3 subunits. Finally, the C-terminal (catalytic) domain catalyzes the transfer of acetyl groups and acetyl-CoA synthesis.[5] In Gammaproteobacteria, 24 copies of E2 form the cubic core of the pyruvate dehydrogenase complex, in which 8 E2 homotrimers are located at the vertices of the cubic core particle.
Dihydrolipoyl dehydrogenase (E3)

The E3 subunit, called the dihydrolipoyl dehydrogenase enzyme, is characterized as a homodimer protein wherein two cysteine residues, engaged in disulfide bonding, and the FAD cofactor in the active site facilitate its main purpose as an oxidizing catalyst. One example of E3 structure, found in Pseudomonas putida, is formed such that each individual homodimer subunit contains two binding domains responsible for FAD binding and NAD binding, as well as a central domain and an interface domain.[6][7]
Dihydrolipoyl dehydrogenase Binding protein (E3BP)
An auxiliary protein unique to most eukaryotes is the E3 binding protein (E3BP), which serves to bind the E3 subunit to the PDC complex. In the case of human E3BP, hydrophobic proline and leucine residues in the BP interact with the surface recognition site formed by the binding of two identical E3 monomers.[8]
Mechanism
This article needs additional citations for verification. (December 2023) |
| Enzymes | Abbrev. | Cofactors | # subunits prokaryotes | # subunits eukaryotes |
|---|---|---|---|---|
| pyruvate dehydrogenase (EC 1.2.4.1) |
E1 | TPP (thiamine pyrophosphate), Mg2+ | 32 | 30 |
| dihydrolipoyl transacetylase (EC 2.3.1.12) |
E2 | alpha-lipoic acid (lipoate) |
24 | 60 |
| dihydrolipoyl dehydrogenase (EC 1.8.1.4) |
E3 | FAD |
16 | 12 |

Pyruvate dehydrogenase (E1)
Initially, pyruvate and thiamine pyrophosphate (TPP or vitamin B1) are bound by pyruvate dehydrogenase subunits.[1] The thiazolium ring of TPP is in a zwitterionic form, and the anionic C2 carbon performs a nucleophilic attack on the C2 (ketone) carbonyl of pyruvate. The resulting intermediate undergoes decarboxylation to produce an acyl anion equivalent (see cyanohydrin or aldehyde-dithiane umpolung chemistry, as well as benzoin condensation). This anion attacks S1 of an oxidized lipoate species that is attached to a lysine residue. In a ring-opening SN2-like mechanism, S2 is displaced as a sulfide or sulfhydryl moiety. Subsequent collapse of the tetrahedral intermediate ejects thiazole, releasing the TPP cofactor and generating a thioacetate on S1 of lipoate. The E1-catalyzed process is the rate-limiting step of the whole pyruvate dehydrogenase complex.
Dihydrolipoyl transacetylase (E2)
At this point, the lipoate-thioester functionality is translocated into the dihydrolipoyl transacetylase (E2) active site,[1] where a transacylation reaction transfers the acetyl from the "swinging arm" of lipoyl to the thiol of coenzyme A. This produces acetyl-CoA, which is released from the enzyme complex and subsequently enters the citric acid cycle. E2 can also be known as lipoamide reductase-transacetylase.
Dihydrolipoyl dehydrogenase (E3)
The dihydrolipoate, covalently bound to a lysine residue of the complex, is then transferred to the dihydrolipoyl dehydrogenase (E3) active site,[1] where it undergoes a flavin-mediated oxidation, similar in chemistry to e.g. thioredoxin reductase. First, FAD oxidizes dihydrolipoate back to its lipoate (disulfide) resting state, producing FADH2. Then, the substrate NAD+ oxidizes FADH2 back to its FAD resting state, producing NADH and H+.
Differences in structure and subunit composition between species
In all organisms, PDC is a large complex composed of multiple copies of the three catalytic subunits E1, E2 and E3. Another common feature of all PDCs is the fact that the subunit E2 forms the core of the complex to which the peripheral subunits E1 and E3 bind. Eukaryotic PDCs contain an additional, non-catalytic subunit in the core termed E3 binding protein (E3BP) (sometimes also "protein X"). In PDCs with a hetero-oligomeric core with multiple copies of E2 and E3BP, E1 exclusively associates with E2, and E3 only binds to E3BP. In contrast, E1 and E3 compete for binding to E2 in bacterial PDCs with a homo-oligomeric E2 core. While the peripheral enzyme E3 is a homodimer in all organisms, the peripheral enzyme E1 is an alpha2beta2 heterotetramerin eukaryotes.
Gram-negative bacteria
In Gram-negative bacteria, e.g. Escherichia coli, PDC consists of a central cubic core made up from 24 molecules of dihydrolipoyl transacetylase (E2). Up to 16 homodimers of pyruvate dehydrogenase (E1) and 8 homodimers of dihydrolipoyl dehydrogenase (E3) bind to the 24 peripheral subunit binding domains (PSBDs) of the E2 24-mer. In Gammaproteobacteria, the specificity of PSBD for binding either E1 or E3 is determined by the oligomeric state of PSBD. In each E2 homotrimer, two of the three PSBDs dimerize. While two E1 homodimers cooperatively bind dimeric PSBD, the remaining, unpaired PSBD specifically interacts with one E3 homodimer. PSBD dimerization thus determines the subunit composition of the pyruvate dehydrogenase complex when fully saturated with the peripheral subunits E1 and E3, which has a stoichiometry of E1:E2:E3 (monomers) = 32:24:16 [9]
Gram-positive bacteria and eukaryotes
In contrast, in Gram-positive bacteria (e.g. Bacillus stearothermophilus) and eukaryotes the central PDC core contains 60 E2 molecules arranged into an icosahedron. This E2 subunit “core” coordinates to 30 subunits of E1 and 12 copies of E3.
Eukaryotes also contain 12 copies of an additional core protein, E3 binding protein (E3BP) which bind the E3 subunits to the E2 core.[10] The exact location of E3BP is not completely clear. Cryo-electron microscopy has established that E3BP binds to each of the icosahedral faces in yeast.[11] However, it has been suggested that it replaces an equivalent number of E2 molecules in the bovine PDC core.
Up to 60 E1 or E3 molecules can associate with the E2 core from Gram-positive bacteria - binding is mutually exclusive. In eukaryotes E1 is specifically bound by E2, while E3 associates with E3BP. It is thought that up to 30 E1 and 6 E3 enzymes are present, although the exact number of molecules can vary in vivo and often reflects the metabolic requirements of the tissue in question.
Regulation
Pyruvate dehydrogenase is inhibited when one or more of the three following ratios are increased: ATP/ADP, NADH/NAD+ and acetyl-CoA/CoA.[citation needed]
In eukaryotes PDC is tightly regulated by its own specific pyruvate dehydrogenase kinase (PDK) and pyruvate dehydrogenase phosphatase (PDP), deactivating and activating it respectively.[12]
- PDK phosphorylates three specific serine residues on E1 with different affinities. Phosphorylation of any one of them (using ATP) renders E1 (and in consequence the entire complex) inactive.[12]
- Dephosphorylation of E1 by PDP reinstates complex activity.[12]
Products of the reaction act as allosteric inhibitors of the PDC, because they activate PDK. Substrates in turn inhibit PDK, reactivating PDC.
During starvation, PDK increases in amount in most tissues, including skeletal muscle, via increased gene transcription. Under the same conditions, the amount of PDP decreases. The resulting inhibition of PDC prevents muscle and other tissues from catabolizing glucose and gluconeogenesis precursors. Metabolism shifts toward fat utilization, while muscle protein breakdown to supply gluconeogenesis precursors is minimized, and available glucose is spared for use by the brain.[citation needed]
Calcium ions have a role in regulation of PDC in muscle tissue, because it activates PDP, stimulating glycolysis on its release into the cytosol - during muscle contraction. Some products of these transcriptions release H2 into the muscles. This can cause calcium ions to decay over time.[citation needed]
Localization of pyruvate decarboxylation
In eukaryotic cells the pyruvate decarboxylation occurs inside the mitochondrial matrix, after transport of the substrate, pyruvate, from the cytosol. The transport of pyruvate into the mitochondria is via the transport protein pyruvate translocase. Pyruvate translocase transports pyruvate in a symport fashion with a proton (across the inner mitochondrial membrane), which may be considered to be a form of secondary active transport, but further confirmation/support may be needed for the usage of "secondary active transport" desciptor here (Note: the pyruvate transportation method via the pyruvate translocase appears to be coupled to a proton gradient according to S. Papa et al., 1971, seemingly matching secondary active transport in definition).[13]
Alternative sources say "transport of pyruvate across the outer mitochondrial membrane appears to be easily accomplished via large non-selective channels such as voltage-dependent anion channels, which enable passive diffusion" and transport across inner mitochondrial membrane is mediated by mitochondrial pyruvate carrier 1 (MPC1) and mitochondrial pyruvate carrier 2 (MPC2).[14]
Upon entry into the mitochondrial matrix, the pyruvate is decarboxylated, producing acetyl-CoA (and carbon dioxide and NADH). This irreversible reaction traps the acetyl-CoA within the mitochondria (the acetyl-CoA can only be transported out of the mitochondrial matrix under conditions of high oxaloacetate via the citrate shuttle, a TCA intermediate that is normally sparse). The carbon dioxide produced by this reaction is nonpolar and small, and can diffuse out of the mitochondria and out of the cell.[citation needed]
In prokaryotes, which have no mitochondria, this reaction is either carried out in the cytosol, or not at all.[citation needed]
Evolutionary history
It was found that pyruvate dehydrogenase enzyme found in the mitochondria of eukaryotic cells closely resembles an enzyme from Geobacillus stearothermophilus, which is a species of gram-positive bacteria. Despite similarities of the pyruvate dehydrogenase complex with gram-positive bacteria, there is little resemblance with those of gram-negative bacteria. Similarities of the quaternary structures between pyruvate dehydrogenase and enzymes in gram-positive bacteria point to a shared evolutionary history which is distinctive from the evolutionary history of corresponding enzymes found in gram-negative bacteria. Through an endosymbiotic event, pyruvate dehydrogenase found in the eukaryotic mitochondria points to ancestral linkages dating back to gram-positive bacteria.[15]
Pyruvate dehydrogenase complexes share many similarities with branched chain 2-oxoacid dehydrogenase (BCOADH), particularly in their substrate specificity for alpha-keto acids. Specifically, BCOADH catalyzes the degradation of amino acids and these enzymes would have been prevalent during the periods on prehistoric Earth dominated by rich amino acid environments. The E2 subunit from pyruvate dehydrogenase evolved from the E2 gene found in BCOADH while both enzymes contain identical E3 subunits due to the presence of only one E3 gene. Since the E1 subunits have a distinctive specificity for particular substrates, the E1 subunits of pyruvate dehydrogenase and BCOADH vary but share genetic similarities. The gram-positive bacteria and cyanobacteria that would later give rise to mitochondria and chloroplast found in eukaryotic cells retained the E1 subunits that are genetically related to those found in the BCOADH enzymes.[16][17]
Clinical relevance
Pyruvate dehydrogenase deficiency (PDCD) can result from mutations in any of the enzymes or cofactors used to build the complex. Its primary clinical finding is lactic acidosis.[18] Such PDCD mutations, leading to subsequent deficiencies in NAD and FAD production, hinder oxidative phosphorylation processes that are key in aerobic respiration. Thus, acetyl-CoA is instead reduced via anaerobic mechanisms into other molecules like lactate, leading to an excess of bodily lactate and associated neurological pathologies.[19]
While pyruvate dehydrogenase deficiency is rare, there are a variety of different genes when mutated or nonfunctional that can induce this deficiency. First, the E1 subunit of pyruvate dehydrogenase contains four different subunits: two alpha subunits designated as E1-alpha and two beta subunits designated as E1-beta. The PDHA1 gene found in the E1-alpha subunits, when mutated, causes 80% of the cases of pyruvate dehydrogenase deficiency because this mutation abridges the E1-alpha protein. Decreased functional E1 alpha prevents pyruvate dehydrogenase from sufficiently binding to pyruvate, thus reducing the activity of the overall complex.[20] When the PDHB gene found in the E1 beta subunit of the complex is mutated, this also leads to pyruvate dehydrogenase deficiency.[21] Likewise, mutations found on other subunits of the complex, like the DLAT gene found on the E2 subunit, the PDHX gene found on the E3 subunit, as well as a mutation on a pyruvate dehydrogenase phosphatase gene, known as PDP1, have all been traced back to pyruvate dehydrogenase deficiency, while their specific contribution to the disease state is unknown.[22][23][24]
See also
References
- ^ a b c d DeBrosse SD, Kerr DS (2016-01-01), Saneto RP, Parikh S, Cohen BH (eds.), "Chapter 12 - Pyruvate Dehydrogenase Complex Deficiency", Mitochondrial Case Studies, Boston: Academic Press, pp. 93–101, doi:10.1016/b978-0-12-800877-5.00012-7, ISBN 978-0-12-800877-5, retrieved 2020-11-16
- ^ J. M. Berg, J. L. Tymoczko, L. Stryer (2007). Biochemistry (6th ed.). Freeman. ISBN 978-0-7167-8724-2.
- ^ Sgrignani J, Chen J, Alimonti A (2018). "How phosphorylation influences E1 subunit pyruvate dehydrogenase: A computational study". Scientific Reports. 8 (14683): 14683. Bibcode:2018NatSR...814683S. doi:10.1038/s41598-018-33048-z. PMC 6168537. PMID 30279533. S2CID 52910721.
- ^ [PBD ID: 2QTC] Kale S, Arjunan P, Furey W, Jordan F (2007). "A dynamic loop at the active center of the Escherichia coli pyruvate dehydrogenase COMPLEX E1 component Modulates SUBSTRATE utilization and CHEMICAL communication with the E2 component". Journal of Biological Chemistry. 282 (38): 28106–28116. doi:10.1074/jbc.m704326200. PMID 17635929. S2CID 25199383.
- ^ Patel MS, Nemeria NS, Furey W, Jordan F (2014). "The pyruvate dehydrogenase complexes: structure-based function and regulation". The Journal of Biological Chemistry. 289 (24): 16615–16623. doi:10.1074/jbc.R114.563148. PMC 4059105. PMID 24798336.
- ^ Billgren ES, Cicchillo RM, Nesbitt NM, Booker SJ (2010). "Lipoic Acid Biosynthesis and Enzymology". Comprehensive Natural Products. 2 (7): 181–212. doi:10.1016/B978-008045382-8.00137-4.
- ^ [PDB ID: 1LVL] Mattevia A, Obmolova G, Sokatch JR, Betzel C, Hol WG (1992). "The refined crystal STRUCTURE of pseudomonas Putida LIPOAMIDE DEHYDROGENASE complexed with NAD+ at 2.45 Å resolution". Proteins: Structure, Function, and Genetics. 13 (4): 336–351. doi:10.1002/prot.340130406. PMID 1325638. S2CID 23288363.
- ^ Ciszak EM, Makal A, Hong YS, Vettaikkorumakankauv AK, Korotchkina LG, Patel MS (2006). "How dihydrolipoamide dehydrogenase-binding protein binds dihydrolipoamide dehydrogenase in the human pyruvate dehydrogenase complex". Journal of Biological Chemistry. 281 (1): 648–655. doi:10.1074/jbc.m507850200. PMID 16263718. S2CID 26797600.
- ^ Meinhold S, Zdanowicz R, Giese C, Glockshuber R (2024). "Dimerization of a 5-kDa domain defines the architecture of the 5-MDa gammaproteobacterial pyruvate dehydrogenase complex". Sci. Adv. 10 (6): eadj6358. Bibcode:2024SciA...10J6358M. doi:10.1126/sciadv.adj6358. PMC 10849603. PMID 38324697. [1]
- ^ Brautigam CA, Wynn RM, Chuang JL, Machius M, Tomchick DR, Chuang DT (2006). "Structural insight into interactions between Dihydrolipoamide Dehydrogenase (E3) and E3 binding protein of Human pyruvate dehydrogenase complex". Structure. 14 (3): 611–621. doi:10.1016/j.str.2006.01.001. PMC 2879633. PMID 16442803.
- ^ Stoops, J.K., Cheng, R.H., Yazdi, M.A., Maeng, C.Y., Schroeter, J.P., Klueppelberg, U., Kolodziej, S.J., Baker, T.S., Reed, L.J. (1997) On the unique structural organization of the Saccharomyces cerevisiae pyruvate dehydrogenase complex. J. Biol. Chem. 272, 5757-5764.
- ^ a b c Pelley JW (2012-01-01), Pelley JW (ed.), "6 - Glycolysis and Pyruvate Oxidation", Elsevier's Integrated Review Biochemistry (Second Edition), Philadelphia: W.B. Saunders, pp. 49–55, doi:10.1016/b978-0-323-07446-9.00006-4, ISBN 978-0-323-07446-9, retrieved 2020-11-16
- ^ Papa S (30 January 1971). "The transport of pyruvate in rat liver mitochondria". FEBS Lett. 12 (5): 285–288. doi:10.1016/0014-5793(71)80200-4. PMID 11945601.
- ^ Rutter J (23 January 2013). "The long and winding road to the mitochondrial pyruvate carrier". Cancer & Metabolism. 1 (1): 6. doi:10.1186/2049-3002-1-6. PMC 3834494. PMID 24280073.
- ^ Henderson CE, Perham RN, Finch JT (May 1979). "Structure and symmetry of B. stearothermophilus pyruvate dehydrogenase multienzyme complex and implications for eucaryote evolution". Cell. 17 (1): 85–93. doi:10.1016/0092-8674(79)90297-6. ISSN 0092-8674. PMID 455461. S2CID 35282258.
- ^ Schreiner ME, Fiur D, Holátko J, Pátek M, Eikmanns BJ (2005-09-01). "E1 Enzyme of the Pyruvate Dehydrogenase Complex in Corynebacterium glutamicum: Molecular Analysis of the Gene and Phylogenetic Aspects". Journal of Bacteriology. 187 (17): 6005–6018. doi:10.1128/jb.187.17.6005-6018.2005. ISSN 0021-9193. PMC 1196148. PMID 16109942.
- ^ Schnarrenberger C, Martin W (2002-02-01). "Evolution of the enzymes of the citric acid cycle and the glyoxylate cycle of higher plants". European Journal of Biochemistry. 269 (3): 868–883. doi:10.1046/j.0014-2956.2001.02722.x. ISSN 0014-2956. PMID 11846788.
- ^ "Pyruvate dehydrogenase deficiency". Genetics Home Reference. Retrieved March 17, 2013.
- ^ Gupta N, Rutledge C (2019). "Pyruvate Dehydrogenase Complex Deficiency: An Unusual Cause of Recurrent Lactic Acidosis in a Paediatric Critical Care Unit". The Journal of Critical Care Medicine. 5 (2): 71–75. doi:10.2478/jccm-2019-0012. PMC 6534940. PMID 31161145.
- ^ Lissens W, De Meirleir L, Seneca S, Liebaers I, Brown GK, Brown RM, Ito M, Naito E, Kuroda Y, Kerr DS, Wexler ID (March 2000). "Mutations in the X-linked pyruvate dehydrogenase (E1) ? subunit gene (PDHA1) in patients with a pyruvate dehydrogenase complex deficiency". Human Mutation. 15 (3): 209–219. doi:10.1002/(sici)1098-1004(200003)15:3<209::aid-humu1>3.0.co;2-k. ISSN 1059-7794. PMID 10679936. S2CID 29926332.
- ^ Okajima K, Korotchkina L, Prasad C, Rupar T, Phillips III J, Ficicioglu C, Hertecant J, Patel M, Kerr D (April 2008). "Mutations of the E1β subunit gene (PDHB) in four families with pyruvate dehydrogenase deficiency". Molecular Genetics and Metabolism. 93 (4): 371–380. doi:10.1016/j.ymgme.2007.10.135. ISSN 1096-7192. PMID 18164639.
- ^ Head RA, Brown RM, Zolkipli Z, Shahdadpuri R, King MD, Clayton PT, Brown GK (2005-07-27). "Clinical and genetic spectrum of pyruvate dehydrogenase deficiency: Dihydrolipoamide acetyltransferase (E2) deficiency". Annals of Neurology. 58 (2): 234–241. doi:10.1002/ana.20550. ISSN 0364-5134. PMID 16049940. S2CID 38264402.
- ^ Pavlu-Pereira H, Silva MJ, Florindo C, Sequeira S, Ferreira AC, Duarte S, Rodrigues AL, Janeiro P, Oliveira A, Gomes D, Bandeira A, Martins E, Gomes R, Soares S, Tavares de Almeida I, Vicente JB, Rivera I (December 2020). "Pyruvate dehydrogenase complex deficiency: updating the clinical, metabolic and mutational landscapes in a cohort of Portuguese patients". Orphanet Journal of Rare Diseases. 15 (1): 298. doi:10.1186/s13023-020-01586-3. PMC 7579914. PMID 33092611.
- ^ Heo HJ, Kim HK, Youm JB, Cho SW, Song IS, Lee SY, Ko TH, Kim N, Ko KS, Rhee BD, Han J (August 2016). "Mitochondrial pyruvate dehydrogenase phosphatase 1 regulates the early differentiation of cardiomyocytes from mouse embryonic stem cells". Experimental & Molecular Medicine. 48 (8): e254. doi:10.1038/emm.2016.70. ISSN 2092-6413. PMC 5007642. PMID 27538372.
External links
- https://web.archive.org/web/20070405211049/http://www.dentistry.leeds.ac.uk/biochem/MBWeb/mb1/part2/krebs.htm#animat1 - animation of the general mechanism of the PDC (link on upper right) at University of Leeds
- Pyruvate+Dehydrogenase+Complex at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
3D structures
- Zhou H, McCarthy B, O'Connor M, Reed J, Stoops K (Dec 2001). "The remarkable structural and functional organization of the eukaryotic pyruvate dehydrogenase complexes". Proceedings of the National Academy of Sciences of the United States of America. 98 (26): 14802–14807. Bibcode:2001PNAS...9814802Z. doi:10.1073/pnas.011597698. ISSN 0027-8424. PMC 64939. PMID 11752427., bovine kidney pyruvate dehydrogenase complex
- Yu X, Hiromasa Y, Tsen H, Stoops K, Roche E, Zhou H (Jan 2008). "Structures of the Human Pyruvate Dehydrogenase Complex Cores: A Highly Conserved Catalytic Center with Flexible N-Terminal Domains". Structure. 16 (1): 104–114. doi:10.1016/j.str.2007.10.024. ISSN 0969-2126. PMC 4807695. PMID 18184588., human full-length and truncated E2 (tE2) cores of PDC, expressed in E. coli
- Articles with short description
- Short description is different from Wikidata
- Articles needing additional references from December 2023
- All articles needing additional references
- All articles with unsourced statements
- Articles with unsourced statements from December 2023
- Commons category link from Wikidata
- EC 1.2.1
- Cellular respiration
- Glycolysis