Key Points
-
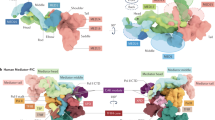
Recent structural advances based on improvements in electron microscopy methodology have enabled the generation of high-resolution structural models of the mediator of RNA polymerase II transcription (Mediator) complex and of the preinitiation complex (PIC) in the presence of Mediator.
-
The module composition of Mediator changes between its recruitment to upstream regulatory regions (enhancers or upstream activating sequences where Mediator is bound to transcription factors) and its action on core promoters together with PIC components.
-
The functional interplay between Mediator and general transcription factors in PIC assembly is closely related to chromatin architecture at promoter regions.
-
Direct contact between Mediator and the nuclear pore-associated transcription-coupled export (TREX2) complex suggests that Mediator functions in gene positioning in the nuclear space.
-
Mediator has been shown to function in the establishment of transcriptional memory, which also involves Mediator interactions with the nuclear pore.
-
Potential therapeutic targeting and modulation of Mediator activity in cancers and in fungal infectious diseases emphasizes the importance of studies of Mediator mechanisms for improving human health.
Abstract
Alterations in the regulation of gene expression are frequently associated with developmental diseases or cancer. Transcription activation is a key phenomenon in the regulation of gene expression. In all eukaryotes, mediator of RNA polymerase II transcription (Mediator), a large complex with modular organization, is generally required for transcription by RNA polymerase II, and it regulates various steps of this process. The main function of Mediator is to transduce signals from the transcription activators bound to enhancer regions to the transcription machinery, which is assembled at promoters as the preinitiation complex (PIC) to control transcription initiation. Recent functional studies of Mediator with the use of structural biology approaches and functional genomics have revealed new insights into Mediator activity and its regulation during transcription initiation, including how Mediator is recruited to transcription regulatory regions and how it interacts and cooperates with PIC components to assist in PIC assembly. Novel roles of Mediator in the control of gene expression have also been revealed by showing its connection to the nuclear pore and linking Mediator to the regulation of gene positioning in the nuclear space. Clear links between Mediator subunits and disease have also encouraged studies to explore targeting of this complex as a potential therapeutic approach in cancer and fungal infections.





Similar content being viewed by others
References
Lee, T. I. & Young, R. A. Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet. 34, 77–137 (2000).
Malik, S. & Roeder, R. G. Transcriptional regulation through Mediator-like coactivators in yeast and metazoan cells. Trends Biochem. Sci. 25, 277–283 (2000).
Myers, L. C. & Kornberg, R. D. Mediator of transcriptional regulation. Annu. Rev. Biochem. 69, 729–749 (2000).
Holstege, F. C. et al. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95, 717–728 (1998).
Soutourina, J., Wydau, S., Ambroise, Y., Boschiero, C. & Werner, M. Direct interaction of RNA polymerase II and mediator required for transcription in vivo. Science 331, 1451–1454 (2011). This study provides in vivo evidence that a direct interaction between Pol II and Mediator is generally required for Pol II recruitment and transcription in yeast.
Thompson, C. M. & Young, R. A. General requirement for RNA polymerase II holoenzymes in vivo. Proc. Natl Acad. Sci. USA 92, 4587–4590 (1995).
Ito, M., Okano, H. J., Darnell, R. B. & Roeder, R. G. The TRAP100 component of the TRAP/Mediator complex is essential in broad transcriptional events and development. EMBO J. 21, 3464–3475 (2002).
Ito, M., Yuan, C. X., Okano, H. J., Darnell, R. B. & Roeder, R. G. Involvement of the TRAP220 component of the TRAP/SMCC coactivator complex in embryonic development and thyroid hormone action. Mol. Cell 5, 683–693 (2000).
Stevens, J. L. et al. Transcription control by E1A and MAP kinase pathway via Sur2 mediator subunit. Science 296, 755–758 (2002).
Tudor, M., Murray, P. J., Onufryk, C., Jaenisch, R. & Young, R. A. Ubiquitous expression and embryonic requirement for RNA polymerase II coactivator subunit Srb7 in mice. Genes Dev. 13, 2365–2368 (1999).
Westerling, T., Kuuluvainen, E. & Makela, T. P. Cdk8 is essential for preimplantation mouse development. Mol. Cell. Biol. 27, 6177–6182 (2007).
Malik, S. & Roeder, R. G. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat. Rev. Genet. 11, 761–772 (2010).
Rhee, H. S. & Pugh, B. F. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature 483, 295–301 (2012). This study uses a precise map of PIC components, including Pol II and GTFs, in the yeast genome to identify TATA-like elements bound by TBP.
Poss, Z. C., Ebmeier, C. C. & Taatjes, D. J. The Mediator complex and transcription regulation. Crit. Rev. Biochem. Mol. Biol. 48, 575–608 (2013).
Cai, G. et al. Mediator head module structure and functional interactions. Nat. Struct. Mol. Biol. 17, 273–279 (2010).
Esnault, C. et al. Mediator-dependent recruitment of TFIIH modules in preinitiation complex. Mol. Cell 31, 337–346 (2008).
Eyboulet, F. et al. Mediator independently orchestrates multiple steps of preinitiation complex assembly in vivo. Nucleic Acids Res. 43, 9214–9231 (2015).
Eychenne, T. et al. Functional interplay between Mediator and TFIIB in preinitiation complex assembly in relation to promoter architecture. Genes Dev. 30, 2119–2132 (2016). On the basis of in vivo, in vitro and in silico approaches, this study reveals a functional interplay between yeast Mediator and TFIIB in close relation to the promoter architecture.
Johnson, K. M., Wang, J., Smallwood, A., Arayata, C. & Carey, M. TFIID and human mediator coactivator complexes assemble cooperatively on promoter DNA. Genes Dev. 16, 1852–1863 (2002).
Lariviere, L. et al. Structure and TBP binding of the Mediator head subcomplex Med8-Med18-Med20. Nat. Struct. Mol. Biol. 13, 895–901 (2006).
Takahashi, H. et al. Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell 146, 92–104 (2011).
Kim, Y. J., Bjorklund, S., Li, Y., Sayre, M. H. & Kornberg, R. D. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 77, 599–608 (1994).
Yudkovsky, N., Ranish, J. A. & Hahn, S. A transcription reinitiation intermediate that is stabilized by activator. Nature 408, 225–229 (2000).
Allen, B. L. & Taatjes, D. J. The Mediator complex: a central integrator of transcription. Nat. Rev. Mol. Cell Biol. 16, 155–166 (2015).
Bourbon, H. M. Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res. 36, 3993–4008 (2008).
Kornberg, R. D. Mediator and the mechanism of transcriptional activation. Trends Biochem. Sci. 30, 235–239 (2005).
Tsai, K. L. et al. Subunit architecture and functional modular rearrangements of the transcriptional mediator complex. Cell 157, 1430–1444 (2014).
Wang, X. et al. Redefining the modular organization of the core Mediator complex. Cell Res. 24, 796–808 (2014).
Cevher, M. A. et al. Reconstitution of active human core Mediator complex reveals a critical role of the MED14 subunit. Nat. Struct. Mol. Biol. 21, 1028–1034 (2014). This work with a reconstituted human Mediator core complex shows that the MED14 subunit is critical for Mediator architecture and function.
Plaschka, C. et al. Architecture of the RNA polymerase II-Mediator core initiation complex. Nature 518, 376–380 (2015). This work provides a cryo-EM model for a recombinant 15-subunit yeast Mediator core within a partially assembled PIC.
Lariviere, L., Seizl, M. & Cramer, P. A structural perspective on Mediator function. Curr. Opin. Cell Biol. 24, 305–313 (2012).
Imasaki, T. et al. Architecture of the Mediator head module. Nature 475, 240–243 (2011).
Lariviere, L. et al. Structure of the Mediator head module. Nature 492, 448–451 (2012).
Robinson, P. J., Bushnell, D. A., Trnka, M. J., Burlingame, A. L. & Kornberg, R. D. Structure of the mediator head module bound to the carboxy-terminal domain of RNA polymerase II. Proc. Natl Acad. Sci. USA 109, 17931–17935 (2012).
Nozawa, K., Schneider, T. R. & Cramer, P. Core Mediator structure at 3.4 A extends model of transcription initiation complex. Nature 545, 248–251 (2017). This work provides the crystal structure for the largest Mediator subcomplex obtained to date at high resolution, the 15-subunit Mediator core from fission yeast.
Robinson, P. J. et al. Molecular architecture of the yeast Mediator complex. eLife 4, e08719 (2015).
Robinson, P. J. et al. Structure of a complete mediator-RNA polymerase II pre-initiation complex. Cell 166, 1411–1422 (2016). In this paper, a cryo-EM model is proposed for a purified yeast Mediator complex lacking the Cdk8 kinase module, which was assembled in vitro with all GTFs and Pol II.
Tsai, K. L. et al. Mediator structure and rearrangements required for holoenzyme formation. Nature 544, 196–201 (2017). This work describes and compares cryo-EM models for fission yeast Mediator alone and in complex with Pol II, revealing conformational changes in Mediator that are associated with Mediator–Pol II interaction.
Sato, S. et al. Role for the MED21-MED7 hinge in assembly of the mediator-RNA polymerase II holoenzyme. J. Biol. Chem. 291, 26886–26898 (2016).
Knuesel, M. T., Meyer, K. D., Bernecky, C. & Taatjes, D. J. The human CDK8 subcomplex is a molecular switch that controls Mediator coactivator function. Genes Dev. 23, 439–451 (2009).
Tsai, K. L. et al. A conserved Mediator-CDK8 kinase module association regulates Mediator-RNA polymerase II interaction. Nat. Struct. Mol. Biol. 20, 611–619 (2013).
Chang, Y. W., Howard, S. C. & Herman, P. K. The Ras/PKA signaling pathway directly targets the Srb9 protein, a component of the general RNA polymerase II transcription apparatus. Mol. Cell 15, 107–116 (2004).
Davis, M. A. et al. The SCF-Fbw7 ubiquitin ligase degrades MED13 and MED13L and regulates CDK8 module association with Mediator. Genes Dev. 27, 151–156 (2013).
Foulds, C. E. et al. Proteomic analysis of coregulators bound to ERalpha on DNA and nucleosomes reveals coregulator dynamics. Mol. Cell 51, 185–199 (2013).
Gonzalez, D. et al. Suppression of Mediator is regulated by Cdk8-dependent Grr1 turnover of the Med3 coactivator. Proc. Natl Acad. Sci. USA 111, 2500–2505 (2014).
Lewicki, M. C., Srikumar, T., Johnson, E. & Raught, B. The S. cerevisiae SUMO stress response is a conjugation-deconjugation cycle that targets the transcription machinery. J. Proteom. 118, 39–48 (2015).
Miller, C. et al. Mediator phosphorylation prevents stress response transcription during non-stress conditions. J. Biol. Chem. 287, 44017–44026 (2012).
van de Peppel, J. et al. Mediator expression profiling epistasis reveals a signal transduction pathway with antagonistic submodules and highly specific downstream targets. Mol. Cell 19, 511–522 (2005).
Mylona, A. et al. Opposing effects of Elk-1 multisite phosphorylation shape its response to ERK activation. Science 354, 233–237 (2016). This study proposes a regulatory mechanism for the fine-tuning of interactions between mammalian Mediator and the ELK1 activator through differential multisite phosphorylation to shape the transcriptional response.
Andrau, J. C. et al. Genome-wide location of the coactivator mediator: Binding without activation and transient Cdk8 interaction on DNA. Mol. Cell 22, 179–192 (2006).
Eyboulet, F. et al. Mediator links transcription and DNA repair by facilitating Rad2/XPG recruitment. Genes Dev. 27, 2549–2562 (2013).
Kagey, M. H. et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467, 430–435 (2010).
Venters, B. J. et al. A comprehensive genomic binding map of gene and chromatin regulatory proteins in Saccharomyces. Mol. Cell 41, 480–492 (2011).
Wong, K. H. & Struhl, K. The Cyc8-Tup1 complex inhibits transcription primarily by masking the activation domain of the recruiting protein. Genes Dev. 25, 2525–2539 (2011).
Ansari, S. A., He, Q. & Morse, R. H. Mediator complex association with constitutively transcribed genes in yeast. Proc. Natl Acad. Sci. USA 106, 16734–16739 (2009).
Kuras, L., Borggrefe, T. & Kornberg, R. D. Association of the Mediator complex with enhancers of active genes. Proc. Natl Acad. Sci. USA 100, 13887–13891 (2003).
Loven, J. et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334 (2013).
Whyte, W. A. et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013).
Jeronimo, C. & Robert, F. Kin28 regulates the transient association of Mediator with core promoters. Nat. Struct. Mol. Biol. 21, 449–455 (2014).
Wong, K. H., Jin, Y. & Struhl, K. TFIIH phosphorylation of the Pol II CTD stimulates mediator dissociation from the preinitiation complex and promoter escape. Mol. Cell 54, 601–612 (2014).
Grunberg, S., Henikoff, S., Hahn, S. & Zentner, G. E. Mediator binding to UASs is broadly uncoupled from transcription and cooperative with TFIID recruitment to promoters. EMBO J. 35, 2435–2446 (2016). This work based on an alternative approach to chromatin immunoprecipitation shows cooperative chromatin binding between budding yeast Mediator and TFIID.
Jeronimo, C. et al. Tail and kinase modules differently regulate core Mediator recruitment and function in vivo. Mol. Cell 64, 455–466 (2016).
Petrenko, N., Jin, Y., Wong, K. H. & Struhl, K. Mediator undergoes a compositional change during transcriptional activation. Mol. Cell 64, 443–454 (2016). References 62 and 63 demonstrate the changes in modular Mediator organization in transcription regulation, presenting a differential role for the tail and kinase modules in Mediator recruitment in budding yeast.
Lai, F. et al. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature 494, 497–501 (2013).
Li, W. et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 498, 516–520 (2013).
Malik, S., Baek, H. J., Wu, W. & Roeder, R. G. Structural and functional characterization of PC2 and RNA polymerase II-associated subpopulations of metazoan Mediator. Mol. Cell. Biol. 25, 2117–2129 (2005).
Pavri, R. et al. PARP-1 determines specificity in a retinoid signaling pathway via direct modulation of mediator. Mol. Cell 18, 83–96 (2005).
Park, S. W. et al. Thyroid hormone-induced juxtaposition of regulatory elements/factors and chromatin remodeling of Crabp1 dependent on MED1/TRAP220. Mol. Cell 19, 643–653 (2005).
Wang, Q., Carroll, J. S. & Brown, M. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol. Cell 19, 631–642 (2005).
Koleske, A. J., Buratowski, S., Nonet, M. & Young, R. A. A novel transcription factor reveals a functional link between the RNA polymerase II CTD and TFIID. Cell 69, 883–894 (1992).
Ranish, J. A., Yudkovsky, N. & Hahn, S. Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev. 13, 49–63 (1999).
Ansari, S. A. et al. Mediator, TATA-binding protein, and RNA polymerase II contribute to low histone occupancy at active gene promoters in yeast. J. Biol. Chem. 289, 14981–14995 (2014).
Kremer, S. B. et al. Role of Mediator in regulating Pol II elongation and nucleosome displacement in Saccharomyces cerevisiae. Genetics 191, 95–106 (2012).
Liu, Z. & Myers, L. C. Med5(Nut1) and Med17(Srb4) are direct targets of mediator histone H4 tail interactions. PLoS ONE 7, e38416 (2012).
Lorch, Y., Beve, J., Gustafsson, C. M., Myers, L. C. & Kornberg, R. D. Mediator-nucleosome interaction. Mol. Cell 6, 197–201 (2000).
Nock, A., Ascano, J. M., Barrero, M. J. & Malik, S. Mediator-regulated transcription through the +1 nucleosome. Mol. Cell 48, 837–848 (2012).
Zhu, X. et al. Histone modifications influence mediator interactions with chromatin. Nucleic Acids Res. 39, 8342–8354 (2011).
Koleske, A. J. & Young, R. A. An RNA polymerase II holoenzyme responsive to activators. Nature 368, 466–469 (1994).
Nonet, M. L. & Young, R. A. Intragenic and extragenic suppressors of mutations in the heptapeptide repeat domain of Saccharomyces cerevisiae RNA polymerase II. Genetics 123, 715–724 (1989).
Thompson, C. M., Koleske, A. J., Chao, D. M. & Young, R. A. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell 73, 1361–1375 (1993).
Reeves, W. M. & Hahn, S. Activator-independent functions of the yeast mediator sin4 complex in preinitiation complex formation and transcription reinitiation. Mol. Cell. Biol. 23, 349–358 (2003).
Tardiff, D. F., Abruzzi, K. C. & Rosbash, M. Protein characterization of Saccharomyces cerevisiae RNA polymerase II after in vivo cross-linking. Proc. Natl Acad. Sci. USA 104, 19948–19953 (2007).
Davis, J. A., Takagi, Y., Kornberg, R. D. & Asturias, F. A. Structure of the yeast RNA polymerase II holoenzyme: Mediator conformation and polymerase interaction. Mol. Cell 10, 409–415 (2002).
Bernecky, C., Grob, P., Ebmeier, C. C., Nogales, E. & Taatjes, D. J. Molecular architecture of the human Mediator-RNA polymerase II-TFIIF assembly. PLoS Biol. 9, e1000603 (2011).
Baek, H. J., Malik, S., Qin, J. & Roeder, R. G. Requirement of TRAP/Mediator for both activator-independent and activator-dependent transcription in conjunction with TFIID-associated TAFIIs. Mol. Cell. Biol. 22, 2842–2852 (2002).
Kuras, L. & Struhl, K. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature 399, 609–613 (1999).
Li, X. Y., Virbasius, A., Zhu, X. & Green, M. R. Enhancement of TBP binding by activators and general transcription factors. Nature 399, 605–609 (1999).
Baek, H. J., Kang, Y. K. & Roeder, R. G. Human Mediator enhances basal transcription by facilitating recruitment of transcription factor IIB during preinitiation complex assembly. J. Biol. Chem. 281, 15172–15181 (2006).
Kang, J. S. et al. The structural and functional organization of the yeast mediator complex. J. Biol. Chem. 276, 42003–42010 (2001).
Jiang, C. & Pugh, B. F. Nucleosome positioning and gene regulation: advances through genomics. Nat. Rev. Genet. 10, 161–172 (2009).
Lai, W. K. M. & Pugh, B. F. Understanding nucleosome dynamics and their links to gene expression and DNA replication. Nat. Rev. Mol. Cell Biol. 18, 548–562 (2017).
Dion, M. F. et al. Dynamics of replication-independent histone turnover in budding yeast. Science 315, 1405–1408 (2007).
Sikorski, T. W. et al. Proteomic analysis demonstrates activator- and chromatin-specific recruitment to promoters. J. Biol. Chem. 287, 35397–35408 (2012).
Nagai, S., Davis, R. E., Mattei, P. J., Eagen, K. P. & Kornberg, R. D. Chromatin potentiates transcription. Proc. Natl Acad. Sci. USA 114, 1536–1541 (2017).
Guglielmi, B., Soutourina, J., Esnault, C. & Werner, M. TFIIS elongation factor and Mediator act in conjunction during transcription initiation in vivo. Proc. Natl Acad. Sci. USA 104, 16062–16067 (2007).
Kim, B. et al. The transcription elongation factor TFIIS is a component of RNA polymerase II preinitiation complexes. Proc. Natl Acad. Sci. USA 104, 16068–16073 (2007).
Hsin, J. P. & Manley, J. L. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 26, 2119–2137 (2012).
Ben-Yishay, R., Ashkenazy, A. J. & Shav-Tal, Y. Dynamic encounters of genes and transcripts with the nuclear pore. Trends Genet. 32, 419–431 (2016).
Bentley, D. L. Coupling mRNA processing with transcription in time and space. Nat. Rev. Genet. 15, 163–175 (2014).
Jani, D., Valkov, E. & Stewart, M. Structural basis for binding the TREX2 complex to nuclear pores, GAL1 localisation and mRNA export. Nucleic Acids Res. 42, 6686–6697 (2014).
Schneider, M. et al. The nuclear pore-associated TREX-2 complex employs mediator to regulate gene expression. Cell 162, 1016–1028 (2015). This study suggests that yeast Mediator is involved in nuclear gene positioning and regulation of transcription via direct contact with the nuclear-pore-associated TREX2 complex.
Hsieh, T. H. et al. Mapping nucleosome resolution chromosome folding in yeast by Micro-C. Cell 162, 108–119 (2015).
D'Urso, A. & Brickner, J. H. Mechanisms of epigenetic memory. Trends Genet. 30, 230–236 (2014).
D'Urso, A. & Brickner, J. H. Epigenetic transcriptional memory. Curr. Genet. 63, 435–439 (2016).
D'Urso, A. et al. Set1/COMPASS and Mediator are repurposed to promote epigenetic transcriptional memory. eLife 5, e16691 (2016). This study proposes a mechanism for Mediator in transcriptional memory that is related to the association of the nuclear pore with corresponding genes.
Elmlund, H. et al. The cyclin-dependent kinase 8 module sterically blocks Mediator interactions with RNA polymerase II. Proc. Natl Acad. Sci. USA 103, 15788–15793 (2006).
Naar, A. M., Taatjes, D. J., Zhai, W., Nogales, E. & Tjian, R. Human CRSP interacts with RNA polymerase II CTD and adopts a specific CTD-bound conformation. Genes Dev. 16, 1339–1344 (2002).
Brickner, D. G. et al. H2A. Z-Mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 5, e81 (2007).
Light, W. H., Brickner, D. G., Brand, V. R. & Brickner, J. H. Interaction of a DNA zip code with the nuclear pore complex promotes H2A. Z incorporation and INO1 transcriptional memory. Mol. Cell 40, 112–125 (2010).
Schiano, C. et al. Involvement of Mediator complex in malignancy. Biochim. Biophys. Acta 1845, 66–83 (2014).
Spaeth, J. M., Kim, N. H. & Boyer, T. G. Mediator and human disease. Semin. Cell Dev. Biol. 22, 776–787 (2011).
Pelish, H. E. et al. Mediator kinase inhibition further activates super-enhancer-associated genes in AML. Nature 526, 273–276 (2015). This work proposes an inhibition of mammalian Mediator kinase as a potential therapeutic for targeting Mediator in cancers.
Nishikawa, J. L. et al. Inhibiting fungal multidrug resistance by disrupting an activator-Mediator interaction. Nature 530, 485–489 (2016). This study proposes that disrupting activator–Mediator contacts could be used for therapeutic targeting of fungal infectious diseases.
Firestein, R. et al. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature 455, 547–551 (2008).
Hnisz, D. et al. Super-enhancers in the control of cell identity and disease. Cell 155, 934–947 (2013).
Poss, Z. C. et al. Identification of Mediator kinase substrates in human cells using cortistatin A and quantitative phosphoproteomics. Cell Rep. 15, 436–450 (2016).
Brzovic, P. S. et al. The acidic transcription activator Gcn4 binds the mediator subunit Gal11/Med15 using a simple protein interface forming a fuzzy complex. Mol. Cell 44, 942–953 (2011).
Thakur, J. K. et al. A nuclear receptor-like pathway regulating multidrug resistance in fungi. Nature 452, 604–609 (2008).
Warfield, L., Tuttle, L. M., Pacheco, D., Klevit, R. E. & Hahn, S. A sequence-specific transcription activator motif and powerful synthetic variants that bind Mediator using a fuzzy protein interface. Proc. Natl Acad. Sci. USA 111, E3506–E3513 (2014).
Malik, S., Wallberg, A. E., Kang, Y. K. & Roeder, R. G. TRAP/SMCC/Mediator-dependent transcriptional activation from DNA and chromatin templates by orphan nuclear receptor hepatocyte nuclear factor 4. Mol. Cell. Biol. 22, 5626–5637 (2002).
Wang, G. et al. Mediator requirement for both recruitment and postrecruitment steps in transcription initiation. Mol. Cell 17, 683–694 (2005).
Tantale, K. et al. A single-molecule view of transcription reveals convoys of RNA polymerases and multi-scale bursting. Nat. Commun. 7, 12248 (2016).
Flanagan, P. M., Kelleher, R. J. 3rd, Sayre, M. H., Tschochner, H. & Kornberg, R. D. A mediator required for activation of RNA polymerase II transcription in vitro. Nature 350, 436–438 (1991).
Backstrom, S., Elfving, N., Nilsson, R., Wingsle, G. & Bjorklund, S. Purification of a plant mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol. Cell 26, 717–729 (2007).
Bourbon, H. M. et al. A unified nomenclature for protein subunits of mediator complexes linking transcriptional regulators to RNA polymerase II. Mol. Cell 14, 553–557 (2004).
Nagulapalli, M., Maji, S., Dwivedi, N., Dahiya, P. & Thakur, J. K. Evolution of disorder in Mediator complex and its functional relevance. Nucleic Acids Res. 44, 1591–1612 (2016).
Toth-Petroczy, A. et al. Malleable machines in transcription regulation: the mediator complex. PLoS Comput. Biol. 4, e1000243 (2008).
Banani, S. F., Lee, H. O., Hyman, A. A. & Rosen, M. K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 (2017).
Hnisz, D., Shrinivas, K., Young, R. A., Chakraborty, A. K. & Sharp, P. A. A. Phase separation model for transcriptional control. Cell 169, 13–23 (2017).
Lariviere, L. et al. Model of the Mediator middle module based on protein cross-linking. Nucleic Acids Res. 41, 9266–9273 (2013).
Pereira-Leal, J. B., Levy, E. D., Kamp, C. & Teichmann, S. A. Evolution of protein complexes by duplication of homomeric interactions. Genome Biol. 8, R51 (2007).
Seizl, M., Lariviere, L., Pfaffeneder, T., Wenzeck, L. & Cramer, P. Mediator head subcomplex Med11/22 contains a common helix bundle building block with a specific function in transcription initiation complex stabilization. Nucleic Acids Res. 39, 6291–6304 (2011).
Risheg, H. et al. A recurrent mutation in MED12 leading to R961W causes Opitz-Kaveggia syndrome. Nat. Genet. 39, 451–453 (2007).
Schwartz, C. E. et al. The original Lujan syndrome family has a novel missense mutation (p. N1007S) in the MED12 gene. J. Med. Genet. 44, 472–477 (2007).
Kaufmann, R. et al. Infantile cerebral and cerebellar atrophy is associated with a mutation in the MED17 subunit of the transcription preinitiation mediator complex. Am. J. Hum. Genet. 87, 667–670 (2010).
Hashimoto, S. et al. MED23 mutation links intellectual disability to dysregulation of immediate early gene expression. Science 333, 1161–1163 (2011).
Kuuselo, R., Savinainen, K., Sandstrom, S., Autio, R. & Kallioniemi, A. MED29, a component of the mediator complex, possesses both oncogenic and tumor suppressive characteristics in pancreatic cancer. Int. J. Cancer 129, 2553–2565 (2011).
Vijayvargia, R., May, M. S. & Fondell, J. D. A coregulatory role for the mediator complex in prostate cancer cell proliferation and gene expression. Cancer Res. 67, 4034–4041 (2007).
Li, L. H., He, J., Hua, D., Guo, Z. J. & Gao, Q. Lentivirus-mediated inhibition of Med19 suppresses growth of breast cancer cells in vitro. Cancer Chemother. Pharmacol. 68, 207–215 (2011).
Boyer, T. G., Martin, M. E., Lees, E., Ricciardi, R. P. & Berk, A. J. Mammalian Srb/Mediator complex is targeted by adenovirus E1A protein. Nature 399, 276–279 (1999).
Brass, A. L. et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science 319, 921–926 (2008).
Bushman, F. D. et al. Host cell factors in HIV replication: meta-analysis of genome-wide studies. PLoS Pathog. 5, e1000437 (2009).
Gwack, Y. et al. Principal role of TRAP/mediator and SWI/SNF complexes in Kaposi's sarcoma-associated herpesvirus RTA-mediated lytic reactivation. Mol. Cell. Biol. 23, 2055–2067 (2003).
Milbradt, A. G. et al. Structure of the VP16 transactivator target in the Mediator. Nat. Struct. Mol. Biol. 18, 410–415 (2011).
Mittler, G. et al. A novel docking site on Mediator is critical for activation by VP16 in mammalian cells. EMBO J. 22, 6494–6504 (2003).
Vojnic, E. et al. Structure and VP16 binding of the Mediator Med25 activator interaction domain. Nat. Struct. Mol. Biol. 18, 404–409 (2011).
Malik, S. Eukaryotic Transcription regulation: getting to the heart of the matter: commentary on Mediator architecture and RNA polymerase II function by Plaschka et al. J. Mol. Biol. 428, 2575–2580 (2016).
Acknowledgements
The author apologizes to the colleagues whose work could not be cited owing to space limitations. The work of J.S. is funded by the Agence Nationale de la Recherche (ANR-14-CE10-0012-01) and the Fondation ARC (grant nos SL220130607079 and PGA1 RF20170205342).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The author declares no competing financial interests.
Supplementary information
Supplementary information S1 (table)
Recent structural models of Mediator complex (PDF 131 kb)
Glossary
- Specific transcription factors
-
Also known as sequence-specific DNA-binding factors (which serve as activators or repressors) that bind to specific DNA sequences in regulatory regions and are essential for regulated transcription.
- Co-regulators
-
Multisubunit protein complexes (which serve as co-activators or co-repressors) that interact with transcription factors and participate in transcription regulation. This group includes complexes that directly influence the basal assembly of transcriptional machinery as well as chromatin modifiers or remodellers that act on a chromatin structure.
- General transcription factors
-
(GTFs). Proteins or protein complexes that are essential for promoter recognition by RNA polymerase II (Pol II), its function and recruitment. This class of proteins were identified as essential components for basal transcription from a specific promoter from in vitro Pol II transcription assays.
- Preinitiation complex
-
(PIC). A large protein complex that assembles on core promoters to position the RNA polymerase. The main components of the PIC of RNA polymerase II (Pol II) are six general transcription factors, Pol II and Mediator.
- Core promoters
-
Minimal DNA sequences close to the transcription start site (TSS) that are sufficient to direct accurate transcription initiation. TATA-elements represent one of the core-promoter elements.
- TATA-elements
-
Consensus sequences (such as TATA-boxes) or sequences presenting some mismatches from the consensus (such as TATA-like elements) in promoters that are bound by TATA-box binding protein.
- Pol II carboxy-terminal domain
-
(CTD) A domain of the largest RNA polymerase II (Pol II) subunit that is important for various steps of the transcription cycle and RNA processing and contains multiple repeats of the heptapeptide sequence YSPTSPS, which are subject to phosphorylation (at Ser2, Ser5 and Ser7) and other post-translational modifications.
- Activating RNAs
-
Long non-coding RNAs with enhancer-like function that interact with Mediator and activate their neighbouring genes.
- Enhancer RNAs
-
Non-coding RNAs that are transcribed from enhancers and contribute to gene looping between enhancers and promoters and thus to gene transcription regulation.
Rights and permissions
About this article
Cite this article
Soutourina, J. Transcription regulation by the Mediator complex. Nat Rev Mol Cell Biol 19, 262–274 (2018). https://doi.org/10.1038/nrm.2017.115
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm.2017.115
- Springer Nature Limited
This article is cited by
-
RhMED15a-like, a subunit of the Mediator complex, is involved in the drought stress response in Rosa hybrida
BMC Plant Biology (2024)
-
Comparative analysis of retroviral Gag-host cell interactions: focus on the nuclear interactome
Retrovirology (2024)
-
Mediator complex subunit 1 promotes oral squamous cell carcinoma progression by activating MMP9 transcription and suppressing CD8+ T cell antitumor immunity
Journal of Experimental & Clinical Cancer Research (2024)
-
5′UTR G-quadruplex structure enhances translation in size dependent manner
Nature Communications (2024)
-
MED15 is upregulated by HIF-2α and promotes proliferation and metastasis in clear cell renal cell carcinoma via activation of SREBP-dependent fatty acid synthesis
Cell Death Discovery (2024)