Host–parasite coevolution
| Part of a series on |
| Evolutionary biology |
|---|
 |
Host–parasite coevolution is a special case of coevolution, where a host and a parasite continually adapt to each other. This can create an evolutionary arms race between them. A more benign possibility is of an evolutionary trade-off between transmission and virulence in the parasite, as if it kills its host too quickly, the parasite will not be able to reproduce either. Another theory, the Red Queen hypothesis, proposes that since both host and parasite have to keep on evolving to keep up with each other, and since sexual reproduction continually creates new combinations of genes, parasitism favours sexual reproduction in the host.
The genetic changes involved are changes in frequencies of alleles, variant forms of individual genes, within populations. These are determined by three main types of selection dynamics: negative frequency-dependent selection when a rare allele has a selective advantage; heterozygote advantage; and directional selection near an advantageous allele. A possible result is a geographic mosaic in a parasitised population, as both host and parasite adapt to environmental conditions that vary in space and time.
Host–parasite coevolution is common in the wild, in humans, in domesticated animals, and in crop plants. Major diseases such as malaria, AIDS and influenza are caused by coevolving parasites.
Model systems for the study of host–parasite coevolution include the nematode Caenorhabditis elegans with the bacterium Bacillus thuringiensis; the crustacean Daphnia and its numerous parasites; and the bacterium Escherichia coli and the mammals (including humans) whose intestines it inhabits.
Overview
[edit]Hosts and parasites exert reciprocal selective pressures on each other, which may lead to rapid reciprocal adaptation. For organisms with short generation times, host–parasite coevolution can be observed in comparatively small time periods, making it possible to study evolutionary change in real-time under both field and laboratory conditions. These interactions may thus serve as a counter-example to the common notion that evolution can only be detected across extended time.[1]
The dynamics of these interactions are summarized in the Red Queen hypothesis, namely that both host and parasite have to change continuously to keep up with each other's adaptations.[2]
Host–parasite coevolution is ubiquitous and of potential importance to all living organisms, including humans, domesticated animals and crops. Major diseases such as malaria, AIDS and influenza are caused by coevolving parasites. Better understanding of coevolutionary adaptations between parasite attack strategies and host immune systems may assist in the development of novel medications and vaccines.[1]
Selection dynamics
[edit]Host–parasite coevolution is characterized by reciprocal genetic change and thus changes in allele frequencies within populations. These changes may be determined by three main types of selection dynamics.[3][1]
Negative frequency-dependent selection
[edit]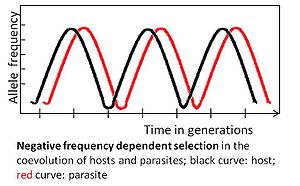
An allele is subject to negative frequency-dependent selection if a rare allelic variant has a selective advantage. For example, the parasite should adapt to the most common host genotype, because it can then infect a large number of hosts. In turn, a rare host genotype may then be favored by selection, its frequency will increase and eventually it becomes common. Subsequently, the parasite should adapt to the former infrequent genotype.[3][4]
Coevolution determined by negative frequency-dependent selection is rapid, potentially occurring across few generations.[3] It maintains high genetic diversity by favoring uncommon alleles. This selection mode is expected for multicellular hosts, because adaptations can occur without the need for novel advantageous mutations, which are less likely to be frequent in these hosts because of relatively small population sizes and relatively long generation times.[3]
Overdominant selection
[edit]Overdominance occurs if the heterozygote phenotype has a fitness advantage over both homozygotes (heterozygote advantage, causing heterosis).[5][6] One example is sickle cell anemia. It is due to a mutation in the hemoglobin gene leading to sickle shape formation of red blood cells, causing clotting in blood vessels, restricted blood flow, and reduced oxygen transport. At the same time, the mutation confers resistance to malaria, caused by Plasmodium parasites, which are passed off in red blood cells after transmission to humans by mosquitoes. Hence, homozygote and heterozygote genotypes for the sickle-cell disease allele show malaria resistance, while the homozygote suffers from severe disease phenotype. The alternative homozygote, which does not carry the sickle cell disease allele, is susceptible to infection by Plasmodium. As a consequence, the heterozygote genotype is selectively favored in areas with a high incidence of malaria.[7][8]
Directional selection
[edit]
If an allele provides a fitness benefit, its frequency increases within a population – selection is directional or positive. A selective sweep is a form of directional selection where the increase in frequency eventually leads to the fixation of the advantageous allele and other alleles near it. The process is considered to be slower than negative frequency-dependent selection. It may produce an "arms race", consisting of the repeated origin and fixation of new virulence traits in the parasite and new defence traits in the host.[1]
This mode of selection is likely to occur in interactions between unicellular organisms and viruses due to large population sizes, short generation times, often haploid genomes and horizontal gene transfer, which increase the probability of beneficial mutations arising and spreading through populations.[3]
Theories
[edit]Geographic mosaic theory of coevolution
[edit]John N. Thompson's geographic mosaic theory of coevolution hypothesizes spatially divergent coevolutionary selection, producing genetic differentiation across populations.[9] The model assumes three elements that jointly fuel coevolution:[10][11][12]

1) a selection mosaic among populations
- Natural selection on interactions between species differs among populations. Thus genotype-by-genotype-by-environment (G x G x E) interactions affect fitness of the antagonists. In other words, the specific environmental conditions determine how any genotype of one species influences the fitness of another species.[10]
2) coevolutionary hotspots
- Coevolutionary hotspots are communities in which selection on the interaction is truly reciprocal. These hotspots are intermixed with so-called coldspots in which only one or neither species adapts to the antagonist.[10]
3) geographic mixing of traits
- Between the communities/regions there is a continuous "mixing" of traits by gene flow, random genetic drift, population extinction, or mutation. This remixing determines the exact dynamics of the geographic mosaic by shifting the spatial distributions of potentially coevolving alleles and traits.[10]
These processes have been intensively studied in a plant, Plantago lanceolata, and its parasite the powdery mildew Podosphaera plantaginis in Åland in south-western Finland.[13] The mildew obtains nutrients from its host, a perennial herb, by sending feeding roots into the plant. There are more than 3000 host populations in this region, where both populations can evolve freely, in the absence of human-imposed selection, in a heterogeneous landscape. Both partners can reproduce asexually and sexually. The mildew tends to become locally extinct in winter, and causes local epidemics in summer. The mildew's success at overwintering, and the intensity of host-pathogen encounters in summer, strongly vary geographically. The system has spatially divergent coevolutionary dynamics across two metapopulations as predicted by the mosaic theory.[14][9]
Red Queen hypothesis
[edit]
The Red Queen hypothesis states that both host and parasite have to change continuously to keep up with each other's adaptations, like the description in Lewis Carroll's fiction.[2] The mathematical evolutionary biologist W. D. Hamilton further proposed that since sexual reproduction continually creates new combinations of genes, some of which may be advantageous, parasitism favours sexual reproduction in the host.[15][16]
The New Zealand freshwater snail Potamopyrgus antipodarum and its different trematode parasites represent a rather special model system. Populations of P. antipodarum consist of asexual clones and sexual individuals and therefore can be used to study the evolution and advantages of sexual reproduction. There is a high correlation between the presence of parasites and the frequency of sexual individuals within the different populations. This result is consistent with the favouring of sexual reproduction proposed in the Red Queen hypothesis.[17]
Trade-off between transmission and virulence
[edit]
Tribolium castaneum, the red flour beetle, is a host for the microsporidian Nosema whitei. This parasitoid kills its host for transmission, so the host's lifespan is important for the parasite's success. In turn, parasite fitness most likely depends on a trade-off between transmission (spore load) and virulence.[18] A higher virulence would increase the potential for the production of more offspring, but a higher spore load would affect the host's lifespan and therefore the transmission rate. This trade-off is supported by coevolutionary experiments, which revealed the decrease of virulence, a constant transmission potential and an increase in the host's lifespan over a period of time.[18] Further experiments demonstrated a higher recombination rate in the host during coevolutionary interactions, which may be selectively advantageous because it should increase diversity of host genotypes.[19]
Resources are generally limited. Therefore, favouring one trait, like virulence or immunity, limits other life-history traits, such as reproductive rate. Moreover, genes are often pleiotropic, having multiple effects. Thus, a change in a pleiotropic immunity or virulence gene can automatically affect other traits. There is thus a trade-off between benefits and costs of the adaptive changes that may prevent the host population from becoming fully resistant or the parasite population from being highly pathogenic. The costs of gene pleiotropy have been investigated in coevolving Escherichia coli and bacteriophages. To inject their genetic material, phages need to bind to a specific bacterial cell surface receptor. The bacterium may prevent injection by altering the relevant binding site, e.g. in response to point mutations or deletion of the receptor. However, these receptors have important functions in bacterial metabolism. Their loss would thus decrease fitness (i.e. population growth rate). As a consequence, there is a trade-off between the advantages and disadvantages of a mutated receptor, leading to polymorphism at this locus.[20]
Model systems for research
[edit]
The nematode Caenorhabditis elegans and the bacterium Bacillus thuringiensis have become established as a model system for studying host–parasite coevolution. Laboratory evolution experiments have provided evidence for many of the basic predictions about these coevolutionary interactions, including reciprocal genetic change, increases in the rate of evolution, and increased genetic diversity.[21]

The crustacean Daphnia and its numerous parasites have become one of the main model systems for studying coevolution. The host can be asexual as well as sexual (induced by changes in the external environment), so sexual reproduction can be stimulated in the laboratory.[3] Decades of coevolution between Daphnia magna and the bacterium Pasteuria ramosa have been reconstructed, reanimating resting stages of both species from laminated pond sediments and exposing hosts from each layer to parasites from the past, the same and the future layers. The study demonstrated that parasites were on average most infective with their contemporary hosts,[22] consistent with negative frequency-dependent selection.[23]

Escherichia coli, a Gram-negative proteobacterium, is a common model in biological research, for which comprehensive data on various aspects of its life-history is available. It has been used extensively for evolution experiments, including those related to coevolution with phages.[20] These studies revealed – among others – that coevolutionary adaptation may be influenced by pleiotropic effects of the involved genes. In particular, binding of the bacteriophage to an E. coli surface receptor is the crucial step in the virus infection cycle. A mutation in the receptor's binding site may cause resistance. Such mutations often show pleiotropic effects and may cause a cost of resistance. In the presence of phages, such pleiotropy may lead to polymorphisms in the bacterial population and thus enhance biodiversity in the community.[20]
Another model system consists of the plant- and animal-colonizing bacterium Pseudomonas and its bacteriophages. This system provided new insights into the dynamics of coevolutionary change. It demonstrated that coevolution may proceed via recurrent selective sweeps, favouring generalists for both partners.[24][25] Furthermore, coevolution with phages may promote allopatric diversity, potentially enhancing biodiversity and possibly speciation.[26] Host–parasite coevolution may also affect the underlying genetics, for example by favouring increased mutation rates in the host.[27]
Tropical tree and liana interactions have also been the subject of study. Here lianas have been viewed as hyper-diverse generalist macro-parasites that affect host survival by parasitising on the host's structural support for access to canopy light, while usurping resources that would otherwise be available to their host. Host trees have widely varying levels of tolerance to infestation of their crowns by lianas.[28][29]
See also
[edit]References
[edit]- ^ a b c d Woolhouse, M. E. J.; Webster, J. P.; Domingo, E.; Charlesworth, B.; Levin, B. R. (December 2002). "Biological and biomedical implications of the coevolution of pathogens and their hosts" (PDF). Nature Genetics. 32 (4): 569–77. doi:10.1038/ng1202-569. hdl:1842/689. PMID 12457190. S2CID 33145462.
- ^ a b Rabajante, J.; et al. (2016). "Host–parasite Red Queen dynamics with phase-locked rare genotypes". Science Advances. 2 (3): e1501548. Bibcode:2016SciA....2E1548R. doi:10.1126/sciadv.1501548. PMC 4783124. PMID 26973878.
- ^ a b c d e f Ebert, D. (2008). "Host–parasite coevolution: Insights from the Daphnia–parasite model system". Current Opinion in Microbiology. 11 (3): 290–301. doi:10.1016/j.mib.2008.05.012. PMID 18556238.
- ^ Rabajante, J.; et al. (2015). "Red Queen dynamics in multi-host and multi-parasite interaction system". Scientific Reports. 5: 10004. Bibcode:2015NatSR...510004R. doi:10.1038/srep10004. PMC 4405699. PMID 25899168.
- ^ Charlesworth, D.; Willis, J. H. (November 2009). "The genetics of inbreeding depression". Nature Reviews Genetics. 10 (11): 783–796. doi:10.1038/nrg2664. PMID 19834483. S2CID 771357.
- ^ Carr, D. E.; Dudash, M. R. (June 2003). "Recent approaches into the genetic basis of inbreeding depression in plants". Philosophical Transactions of the Royal Society of London B: Biological Sciences. 358 (1434): 1071–1084. doi:10.1098/rstb.2003.1295. PMC 1693197. PMID 12831473.
- ^ "What Is Sickle Cell Disease?". National Heart, Lung, and Blood Institute. June 12, 2015. Archived from the original on 6 March 2016. Retrieved 8 March 2016.
- ^ Wellems, T. E.; Hayton, K.; Fairhurst, R. M. (September 2009). "The impact of malaria parasitism: from corpuscles to communities". Journal of Clinical Investigation. 119 (9): 2496–505. doi:10.1172/JCI38307. PMC 2735907. PMID 19729847.
- ^ a b Laine, Anna-Liisa (July 2009). "Role of coevolution in generating biological diversity - spatially divergent selection trajectories". Journal of Experimental Botany. 60 (11): 2957–2970. doi:10.1093/jxb/erp168. PMID 19528527.
- ^ a b c d "The John N Thompson Lab". University of California Santa Cruz. Retrieved 7 January 2018.
- ^ Thompson, John N. (2005). The Geographic Mosaic of Coevolution (Interspecific Interactions). University of Chicago Press. ISBN 978-0-226-79762-5.
- ^ Thompson, John N. (1999). "Specific Hypotheses on the Geographic Mosaic of Coevolution". The American Naturalist. 153: S1–S14. doi:10.1086/303208. S2CID 11656923.
- ^ a b Soubeyrand, S.; Laine, Anna-Liisa; Hanski, I.; Penttinen, A. (2009). "Spatio-temporal structure of host-pathogen interactions in a metapopulation". The American Naturalist. 174 (3): 308–320. doi:10.1086/603624. PMID 19627233. S2CID 38975948.
- ^ Laine, Anna-Liisa (2005). Linking spatial and evolutionary dynamics in a plant-pathogen metapopulation (PDF). Department of Biological and Environmental Sciences, University of Helsinki, Finland (PhD thesis).
- ^ Hamilton, W. D.; Axelrod, R.; Tanese, R. (1990). "Sexual reproduction as an adaptation to resist parasites". Proceedings of the National Academy of Sciences of the USA. 87 (9): 3566–3573. Bibcode:1990PNAS...87.3566H. doi:10.1073/pnas.87.9.3566. PMC 53943. PMID 2185476.
- ^ Hamilton, W. D. (1980). "Sex versus non-sex versus parasite". Oikos. 35 (2): 282–290. doi:10.2307/3544435. JSTOR 3544435.
- ^ Jokela, Jukka; Liveley, Curtis M.; Dydahl, Mark F.; Fox, Jennifer A. (7 May 2003). "Genetic variation in sexual and clonal lineages of a freshwater snail". Biological Journal of the Linnean Society. 79 (1): 165–181. doi:10.1046/j.1095-8312.2003.00181.x.
- ^ a b Bérénos, C.; Schmid-Hempel, P.; Wegner, K. M. (October 2009). "Evolution of host resistance and trade-offs between virulence and transmission potential in an obligately killing parasite". Journal of Evolutionary Biology. 22 (10): 2049–56. doi:10.1111/j.1420-9101.2009.01821.x. PMID 19732263. S2CID 19399783.
- ^ Fischer, O.; Schmid-Hempel, P. (2005). "Selection by parasites may increase host recombination frequency". Biology Letters. 22 (2): 193–195. doi:10.1098/rsbl.2005.0296. PMC 1626206. PMID 17148164.
- ^ a b c Bohannan, B. J. M.; Lenski, R. E. (2000). "Linking genetic change to community evolution: insights from studies of bacteria and bacteriophage". Ecology Letters. 3 (4): 362–77. doi:10.1046/j.1461-0248.2000.00161.x.
- ^ Schulte, R. D.; Makus, C.; Hasert, B.; Michiels, N. K.; Schulenburg, H. (20 April 2010). "Multiple reciprocal adaptations and rapid genetic change upon experimental coevolution of an animal host and its microbial parasite". PNAS. 107 (16): 7359–7364. Bibcode:2010PNAS..107.7359S. doi:10.1073/pnas.1003113107. PMC 2867683. PMID 20368449.
- ^ Decaestecker, E.; Gaba, S.; Raeymaekers, J. A.; Stoks, R.; Van Kerckhoven, L.; Ebert, D.; De Meester, L. (6 December 2007). "Host–parasite 'Red Queen' dynamics archived in pond sediment". Nature. 450 (7171): 870–3. Bibcode:2007Natur.450..870D. doi:10.1038/nature06291. PMID 18004303. S2CID 4320335.
- ^ Gandon, S.; Buckling, A.; Decaestecker, E.; Day, T. (November 2008). "Host–parasite coevolution and patterns of adaptation across time and space". Journal of Evolutionary Biology. 21 (6): 1861–1866. doi:10.1111/j.1420-9101.2008.01598.x. PMID 18717749. S2CID 31381381.
- ^ Buckling, A.; Rainey, P. B. (2002a). "Antagonistic coevolution between a bacterium and a bacteriophage". Proceedings of the Royal Society B: Biological Sciences. 269 (1494): 931–936. doi:10.1098/rspb.2001.1945. PMC 1690980. PMID 12028776.
- ^ Brockhurst, M. A.; Morgan, A. D.; Fenton, A.; Buckling, A. (2007). "Experimental coevolution with bacteria and phage: the Pseudomonas fluorescens model system". Infection, Genetics and Evolution. 7 (4): 547–552. doi:10.1016/j.meegid.2007.01.005. PMID 17320489.
- ^ Buckling, A.; Rainey, P. B. (2002b). "The role of parasites in sympatric and allopatric host diversification". Nature. 420 (6915): 496–499. Bibcode:2002Natur.420..496B. doi:10.1038/nature01164. PMID 12466840. S2CID 4411588.
- ^ Pal, C.; Macia, M. D.; Oliver, A.; Schachar, I.; Buckling, A. (2007). "Coevolution with viruses drives the evolution of bacterial mutation rates". Nature. 450 (7172): 1079–1081. Bibcode:2007Natur.450.1079P. doi:10.1038/nature06350. PMID 18059461. S2CID 4373536.
- ^ Visser, Marco D.; Muller-Landau, Helene C.; Schnitzer, Stefan A.; de Kroon, Hans; Jongejans, Eelke; Wright, S. Joseph; Gibson, David (2018). "A host–parasite model explains variation in liana infestation among co-occurring tree species". Journal of Ecology. 106 (6): 2435–2445. doi:10.1111/1365-2745.12997. hdl:2066/193285.
- ^ Visser, Marco D.; Schnitzer, Stefan A.; Muller-Landau, Helene C.; Jongejans, Eelke; de Kroon, Hans; Comita, Liza S.; Hubbell, Stephen P.; Wright, S. Joseph; Zuidema, Pieter (2018). "Tree species vary widely in their tolerance for liana infestation: A case study of differential host response to generalist parasites". Journal of Ecology. 106 (2): 781–794. doi:10.1111/1365-2745.12815. hdl:2066/176867.

