Actinin
Actinin is a microfilament protein. The functional protein is an anti-parallel dimer, which cross-links the thin filaments in adjacent sarcomeres, and therefore coordinates contractions between sarcomeres in the horizontal axis. Alpha-actinin is a part of the spectrin superfamily. This superfamily is made of spectrin, dystrophin, and their homologous and isoforms. In non-muscle cells, it is found by the actin filaments and at the adhesion sites[1].The lattice like arrangement provides stability to the muscle contractile apparatus.[1] Specifically, it helps bind actin filaments to the cell membrane.[2] There is a binding site at each end of the rod and with bundles of actin filaments.[1]
The non-sarcomeric alpha-actinins, encoded by ACTN1 and ACTN4, are widely expressed. ACTN2 expression is found in both cardiac and skeletal muscle, whereas ACTN3 is limited to the latter. Both ends of the rod-shaped alpha-actinin dimer contain actin-binding domains. Six different proteins are produced from four alpha-actinin encoding genes.These six proteins can further be divided into two different groups: muscle (calcium insensitive) and non-muscle cytoskeletal (calcium sensitive) isoforms.[1]
Evolution
[edit]There is belief that there is a common alpha-actinin like ancestor gene when looking at features in alpha-actinin and spectrin.[3] Examining spectrin repeat sequences provides evidence for a two-stage model describing the evolution of the spectrin superfamily. In looking at their common ancestor, alpha-actinin and spectrin have four homologous repeats.[3] A gene duplication resulted in the emergence of a stable lineage that led to modern alpha-actinin genes. Simultaneously, the other duplicated gene acquired extra repeats through a series of unequal crossing-over events. This made the spectrin subunit ancestor which is an antiparallel homodimer that can crosslink actin filaments.[3] Alpha-actinin 1 (ACTN1) was discovered forty years ago due to it being present in the striated muscle contractile apparatus in large amounts.[4] Alpha-actinin-1 is necessary for the attachment of actin myofilaments to the Z-lines in skeletal muscle cells,[5] and to the dense bodies in smooth muscle cells.[6] Alpha-actinin 2 (ACTN2) is mainly found in cardiac and oxidative muscle fibers. Some of ACTN2 is seen in the brain. Alpha-actinin 3 (ACTN3) is typically found in type II muscle fibers, commonly known as fast twitch muscle fibers.[4]
Structure
[edit]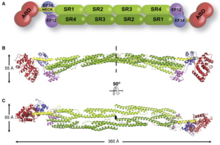
It has a N-terminus which all members of the superfamily have. This is made up of two consecutive calponin homology (CH) where spectrin repeats comes right after it. This allows for the length and flexibility of the actin binding protein to be decided. The actin-filament cross-links involve alpha-actinin, which is a functional anti-parallel dimer.[1] It consists of an actin binding domain (ABD) connected to four spectrin repeats forming the central rod through a flexible neck region. These repeats are 122 amino acid repeats.[8] This is then followed by a C-terminal calmodulin (CaM)-like domain which contain two EF-hand calcium binding motifs.[8][1] This forms the binding site at each end of the protein which results in a rod-shaped molecule and with bundles of actin filaments.[1] The rod shaped appearance is due to the SR region having a cylindrical shape.[7] At each end there is the functional domain (ABD and CaM).[1] The binding of calcium is only present in ACTN1 and ACTN4, while ACTN2 and ACTN3 have lost the ability to bind calcium.[9]
Actin binding domain
[edit]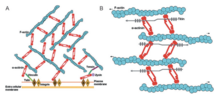
Alpha-actinin and actin are both highly conserved proteins with alpha-actinin being the most conserved in the entire domain in the protein family. This is due to the ABD which binds to type 1 and type 2 CH domains (CH1, and CH2). The CH1-CH2 domain has a hydrophilic stabilizing portion and a hydrophobic part. The core of each CH domain has four helices (A, C, E, and G). Helices C and G are parallel to each other and the N-terminal helix A and E surrounds them. In humans, the crystal structures of ABDs were determined of alpha-actinin 1,3, and 4.[1] The ABD forms a closed conformation. NMR has shown that there are three major ABD. The three sites are the N-terminal of the A helix of CH1, the C-terminal of the G helix of CH1, and to the inter-domain linker bordered by the N-terminal segment of the CH2 domain.[1] These have a high affinity to the actin filaments. However, they must work together to have the highest affinity as CH2 can not bind actin filaments. With this being said, the actin-binding domain is located in the N-terminal region of the alpha-actinin molecule.[8]
Metabolism
[edit]Alpha-actinin 3 (ACTN3) is deficient in around sixteen percent of humans and it plays a significant role in muscle metabolism.[10] This deficiency is due to a premature stop codon polymorphism (R577X).[11] The R577X gene was higher in endurance athletes than in sprint athletes.[12] Among the four mammalian alpha-actinins, ACTN3 stands out as the most highly specialized, primarily expressed in fast glycolytic fibers within skeletal muscle.[11] In humans that have ACTN3, scientists have seen better results in sprinting and power performance in athletes and the general population.[10] Even though this has been found, recent positive selection appears to have influenced the null genotype XX, possibly owing to its emerging role in regulating muscle metabolism, as suggested by the available evidence.[10] The lack of ACTN3 results in a more oxidative pathways of energy being used as glycogen phosphorylase activity is reduced. This lack of ACTN3 does not lead to clear cause for muscle disease[10] but an alteration in muscle function has been seen.[12]
Cancer
[edit]Alpha-actinin 4 (ACTN4) is expressed in non-muscle cells. It is important as it is the link between two tumor components. ACTN4 guides the connection between the actin cytoskeleton within the cell and the integrins that directly interact with the stromal ECM. Additionally, it can sense and respond to externally applied force.[9] This process is crucial for the formation and continuation of breast, colorectal, ovarian, and pancreatic cancer. The explanation as to why ACTN4 contributes to cancer formation is still unknown. In melanoma cancer cells, ACTN4 plays a role in cellular morphology. It changes the cell from a more mesenchymal type cell to an amoeboid type cell by reducing the focal adhesion site.[9] The change in the focal adhesion site is important as focal adhesion sites are critical for the creation assembly of actin stress fibers and the migratory behaviors of cells.[9] Changing from a mesenchymal type cell to an amoeboid type cell allows for a higher rate of invasion through collagen. In mesenchymal type cells, they are reliant on the focal adhesion point and integrin. This allows for their invasion through collagen. Amoeboid type cells lack stress fibers and use high myosin II mediated contraction which allows for it to invade the blebbing mechanism.[9] This process is what scientists are looking at further as this could clarify the significant rise in invasion and metastasis observed as they navigate through the dense stroma of tumors.[9] Mutations in ACTN4 can cause the kidney disease focal segmental glomerulosclerosis (FSGS).[13]
See also
[edit]References
[edit]- ^ a b c d e f g h i j k Sjöblom B, Salmazo A, Djinović-Carugo K (September 2008). "Alpha-actinin structure and regulation". Cellular and Molecular Life Sciences. 65 (17): 2688–2701. doi:10.1007/s00018-008-8080-8. PMC 11131806. PMID 18488141. S2CID 26321210.
- ^ Broderick MJ, Winder SJ (January 2005). "Spectrin, alpha-actinin, and dystrophin". Advances in Protein Chemistry. Fibrous Proteins: Coiled-Coils, Collagen and Elastomers. 70. Academic Press: 203–246. doi:10.1016/S0065-3233(05)70007-3. ISBN 9780120342709. PMID 15837517. Retrieved 2023-11-06.
- ^ a b c Viel A (November 1999). "Alpha-actinin and spectrin structures: an unfolding family story". FEBS Letters. 460 (3): 391–394. doi:10.1016/S0014-5793(99)01372-1. PMID 10556504. S2CID 20832269.
- ^ a b Hsu KS, Kao HY (2013). "Alpha-actinin 4 and tumorigenesis of breast cancer". Vitamins and Hormones. 93: 323–351. doi:10.1016/B978-0-12-416673-8.00005-8. ISBN 9780124166738. PMC 4143506. PMID 23810014.
- ^ Ganong's Review of Medical Physiology, 24th Edition. Lange (Tata McGraw Hill). 2012. p. 100.
- ^ Gabella G (6 December 2012). "Structure of Smooth Muscles". In Szekeres L, Papp JG (eds.). Pharmacology of Smooth Muscle. Springer Science & Business Media. pp. 17–18. ISBN 978-3-642-78920-5.
- ^ a b Ribeiro E, Pinotsis N, Ghisleni A, Salmazo A, Konarev PV, Kostan J, et al. (December 2014). "The structure and regulation of human muscle α-actinin". Cell. 159 (6): 1447–1460. doi:10.1016/j.cell.2014.10.056. PMC 4259493. PMID 25433700.
- ^ a b c Blanchard A, Ohanian V, Critchley D (August 1989). "The structure and function of alpha-actinin". Journal of Muscle Research and Cell Motility. 10 (4): 280–289. doi:10.1007/BF01758424. PMID 2671039. S2CID 22230767.
- ^ a b c d e f Thomas DG, Robinson DN (November 2017). "The fifth sense: Mechanosensory regulation of alpha-actinin-4 and its relevance for cancer metastasis". Seminars in Cell & Developmental Biology. Mechanosensing: from molecules to tissues. 71: 68–74. doi:10.1016/j.semcdb.2017.05.024. PMC 5659936. PMID 28579451.
- ^ a b c d Berman Y, North KN (August 2010). "A gene for speed: the emerging role of alpha-actinin-3 in muscle metabolism". Physiology. 25 (4): 250–259. doi:10.1152/physiol.00008.2010. PMID 20699471.
- ^ a b MacArthur DG, North KN (July 2004). "A gene for speed? The evolution and function of alpha-actinin-3". BioEssays. 26 (7): 786–795. doi:10.1002/bies.20061. PMID 15221860. S2CID 19761762.
- ^ a b Lee FX, Houweling PJ, North KN, Quinlan KG (April 2016). "How does α-actinin-3 deficiency alter muscle function? Mechanistic insights into ACTN3, the 'gene for speed'". Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 1863 (4): 686–693. doi:10.1016/j.bbamcr.2016.01.013. PMID 26802899.
- ^ Polu KR, Pollack MR (25 February 2009). "Focal Segmental Glomerulosclerosis". In Lifton RP, Somlo S, Giebisch GH, Seldin DW (eds.). Genetic Diseases of the Kidney. Academic Press. pp. 117–118. ISBN 978-0-08-092427-4.
External links
[edit]- Actinin at the U.S. National Library of Medicine Medical Subject Headings (MeSH)

