Abstract
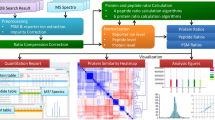
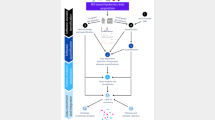
High-resolution mass spectrometry (MS) has become an important tool in the life sciences, contributing to the diagnosis and understanding of human diseases, elucidating biomolecular structural information and characterizing cellular signaling networks. However, the rapid growth in the volume and complexity of MS data makes transparent, accurate and reproducible analysis difficult. We present OpenMS 2.0 (http://www.openms.de), a robust, open-source, cross-platform software specifically designed for the flexible and reproducible analysis of high-throughput MS data. The extensible OpenMS software implements common mass spectrometric data processing tasks through a well-defined application programming interface in C++ and Python and through standardized open data formats. OpenMS additionally provides a set of 185 tools and ready-made workflows for common mass spectrometric data processing tasks, which enable users to perform complex quantitative mass spectrometric analyses with ease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Weisser, H. et al. An automated pipeline for high-throughput label-free quantitative proteomics. J. Proteome Res. 12, 1628–1644 (2013).Extended description of the OpenMS label-free workflow; compares the results to those obtained with other software.
Martens, L. et al. mzML—a community standard for mass spectrometry data. Mol. Cell. Proteomics 10, R110.000133 (2011).
Walzer, M. et al. The mzQuantML data standard for mass spectrometry-based quantitative studies in proteomics. Mol. Cell. Proteomics 12, 2332–2340 (2013).
Griss, J. et al. The mzTab data exchange format: communicating mass-spectrometry-based proteomics and metabolomics experimental results to a wider audience. Mol. Cell. Proteomics 13, 2765–2775 (2014).
Jones, A.R. et al. The mzIdentML data standard for mass spectrometry-based proteomics results. Mol. Cell. Proteomics 11, M111.014381 (2012).
Deutsch, E.W. et al. A guided tour of the trans-proteomic pipeline. Proteomics 10, 1150–1159 (2010).
Chambers, M.C. et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 30, 918–920 (2012).
Sturm, M. et al. OpenMS—an open-source software framework for mass spectrometry. BMC Bioinformatics 9, 163 (2008).Contains the first description of OpenMS as a C++ software library.
Vaudel, M. et al. PeptideShaker enables reanalysis of MS-derived proteomics data sets. Nat. Biotechnol. 33, 22–24 (2015).
Wang, R. et al. PRIDE Inspector: a tool to visualize and validate MS proteomics data. Nat. Biotechnol. 30, 135–137 (2012).
Devil in the details. Nature 470, 305–306 (2011).
Code share. Nature 514, 536 (2014).
Berthold, M.R. et al. KNIME: The Konstanz Information Miner (Springer, 2008).
Goecks, J., Nekrutenko, A. & Taylor, J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 11, R86 (2010).
Döring, A., Weese, D., Rausch, T. & Reinert, K. SeqAn an efficient, generic C. library for sequence analysis. BMC Bioinformatics 9, 11 (2008).
Walzer, M. et al. qcML: an exchange format for quality control metrics from mass spectrometry experiments. Mol. Cell. Proteomics 13, 1905–1913 (2014).
Deutsch, E.W. et al. TraML—a standard format for exchange of selected reaction monitoring transition lists. Mol. Cell. Proteomics 11, R11.015040 (2012).
Röst, H.L., Schmitt, U., Aebersold, R. & Malmström, L. pyOpenMS: a Python-based interface to the OpenMS mass-spectrometry algorithm library. Proteomics 14, 74–77 (2014).
Kiefer, P., Schmitt, U. & Vorholt, J.A. eMZed: an open source framework in Python for rapid and interactive development of LC/MS data analysis workflows. Bioinformatics 29, 963–964 (2013).
Röst, H.L., Rosenberger, G., Aebersold, R. & Malmström, L. Efficient visualization of high-throughput targeted proteomics experiments: TAPIR. Bioinformatics 31, 2415–2417 (2015).
DeLano, W.L. The PyMOL Molecular Graphics System (DeLano Scientific, 2002).
Craig, R. & Beavis, R.C. TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 (2004).
Tyanova, S. et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods http://dx.doi.org/10.1038/nmeth.3901 (2016).
Junker, J. et al. TOPPAS: a graphical workflow editor for the analysis of high-throughput proteomics data. J. Proteome Res. 11, 3914–3920 (2012).
Aiche, S. et al. Workflows for automated downstream data analysis and visualization in large-scale computational mass spectrometry. Proteomics 15, 1443–1447 (2015).Highlights the importance of workflows in the world of MS and discusses open-source software solutions for workflow management.
Kunszt, P. et al. iPortal: the swiss grid proteomics portal: requirements and new features based on experience and usability considerations. Concurr. Comput. 27, 433–445 (2015).
Kessner, D., Chambers, M., Burke, R., Agus, D. & Mallick, P. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics 24, 2534–2536 (2008).
Geer, L.Y. et al. Open mass spectrometry search algorithm. J. Proteome Res. 3, 958–964 (2004).
Kim, S. et al. The generating function of CID, ETD, and CID/ETD pairs of tandem mass spectra: applications to database search. Mol. Cell. Proteomics 9, 2840–2852 (2010).
Käll, L., Canterbury, J.D., Weston, J., Noble, W.S. & MacCoss, M.J. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods 4, 923–925 (2007).
Serang, O., MacCoss, M.J. & Noble, W.S. Efficient marginalization to compute protein posterior probabilities from shotgun mass spectrometry data. J. Proteome Res. 9, 5346–5357 (2010).
Kenar, E. et al. Automated label-free quantification of metabolites from liquid chromatography-mass spectrometry data. Mol. Cell. Proteomics 13, 348–359 (2014).First application of OpenMS to metabolomics.
Kramer, K. et al. Photo-cross-linking and high-resolution mass spectrometry for assignment of RNA-binding sites in RNA-binding proteins. Nat. Methods 11, 1064–1070 (2014).Describes the use of OpenMS to investigate RNA–protein cross-linking.
Röst, H.L. et al. OpenSWATH enables automated, targeted analysis of data-independent acquisition MS data. Nat. Biotechnol. 32, 219–223 (2014).First publication of an automated workflow for targeted analysis of SWATH-MS data, implemented in OpenMS.
Nahnsen, S., Bertsch, A., Rahnenführer, J., Nordheim, A. & Kohlbacher, O. Probabilistic consensus scoring improves tandem mass spectrometry peptide identification. J. Proteome Res. 10, 3332–3343 (2011).
Nilse, L., Sigloch, F.C., Biniossek, M.L. & Schilling, O. Toward improved peptide feature detection in quantitative proteomics using stable isotope labeling. Proteomics Clin. Appl. 9, 706–714 (2015).
Röst, H.L., Schmitt, U., Aebersold, R. & Malmström, L. Fast and efficient XML data access for next-generation mass spectrometry. PLoS One 10, e0125108 (2015).
Bielow, C., Aiche, S., Andreotti, S. & Reinert, K. MSSimulator: simulation of mass spectrometry data. J. Proteome Res. 10, 2922–2929 (2011).
Liu, Y. et al. Quantitative variability of 342 plasma proteins in a human twin population. Mol. Syst. Biol. 11, 786 (2015).
Gillet, L.C. et al. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol. Cell. Proteomics 11, O111.016717 (2012).
Lai, Z.W. et al. Formalin-fixed, paraffin-embedded tissues (FFPE) as a robust source for the profiling of native and protease-generated protein amino termini. Mol. Cell. Proteomics 15, 2203–2213 (2016).
Tholen, S. et al. Contribution of cathepsin L to secretome composition and cleavage pattern of mouse embryonic fibroblasts. Biol. Chem. 392, 961–971 (2011).
Wright, J.C. et al. Improving GENCODE reference gene annotation using a high-stringency proteogenomics workflow. Nat. Commun. 7, 11778 (2016).
Harrow, J. et al. GENCODE: producing a reference annotation for ENCODE. Genome Biol. 7, S4.1–S4.9 (2006).
Petryszak, R. et al. Expression Atlas update—an integrated database of gene and protein expression in humans, animals and plants. Nucleic Acids Res. 44, D746–D752 (2016).
Choi, M. et al. MSstats: an R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics 30, 2524–2526 (2014).
Rosenberger, G., Ludwig, C., Röst, H.L., Aebersold, R. & Malmström, L. aLFQ: an R-package for estimating absolute protein quantities from label-free LC-MS/MS proteomics data. Bioinformatics 30, 2511–2513 (2014).
MacLean, B. et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 (2010).
Acknowledgements
We gratefully acknowledge the contributions of all OpenMS developers as well as of our users who helped to improve the software. This work was funded by ETH (ETH-30 11-2 to H.L.R.), SNSF (P2EZP3_162268 to H.L.R.), the Swiss Federal Commission for Technology and Innovation CTI (13539.1 PFFLI-LS to G.R.), ERC Proteomics v3.0 (233226 to R.A.), the PhosphonetX project of SystemsX.ch (R.A.), the Swiss National Science Foundation (R.A.), the Wellcome Trust (grant WT098051 to the Sanger Institute/H.W., P.G. and J.S.C.), the German Academic Exchange Service (DAAD, grant 57076385 to X.L.), the European Union (FP7, Predict-IV, GA 202222 to K.R. and C.B.), the Deutsche Forschungsgemeinschaft (SCHI 871/5, SCHI 871/6, SCHI 871/8, SCHI 871/9, GR 1748/6, INST 39/900-1, and SFB850-Project B8 to O.S. and L.N.), the European Research Council (ERC-2011-StG 282111-ProteaSys to O.S. and L.N.), DFG (QBiC to S.N., D.W. and O.K.; SFB685 to M.W. and O.K.), the European Union's Seventh Framework Programme (FP7/2007-2013 under EC-GA No. 263215 “MARINA” to M.R., F.A., S.A., K.R. and O.K.), the European Union (PRIME-XS to M.W., S.N. and O.K.), and BMBF (grant 01GI1104A to E.K. and O.K.; grant 01ZX1301F to E.K., J.P., T.S. and O.K.; grant 031A430C to F.A. and O.K.; grant 0315395B to J.V., L.N. and O.K.; grants 031A367 and 031A535A to O.K., K.R., S.A., J.P. and T.S.). We are deeply grateful for the support of the KNIME team, the Proteome Discoverer team and the Compound Discoverer team.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
C.B. is a part-time employee of CodeMS, which operates in the field covered in the Perspective.
Supplementary information
Supplementary Text and Figures
Supplementary Notes 1–3, and Supplementary Protocols 1 and 2 (PDF 5944 kb)
Rights and permissions
About this article
Cite this article
Röst, H., Sachsenberg, T., Aiche, S. et al. OpenMS: a flexible open-source software platform for mass spectrometry data analysis. Nat Methods 13, 741–748 (2016). https://doi.org/10.1038/nmeth.3959
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.3959
This article is cited by
-
ASGR1 deficiency diverts lipids toward adipose tissue but results in liver damage during obesity
Cardiovascular Diabetology (2024)
-
PFΔScreen — an open-source tool for automated PFAS feature prioritization in non-target HRMS data
Analytical and Bioanalytical Chemistry (2024)
-
Decoding cancer insights: recent progress and strategies in proteomics for biomarker discovery
Journal of Proteins and Proteomics (2024)
-
OpenMS 3 enables reproducible analysis of large-scale mass spectrometry data
Nature Methods (2024)
-
Ultra-sensitive isotope probing to quantify activity and substrate assimilation in microbiomes
Microbiome (2023)